New publication in eLife of the Jordan Lab in collaboration with the IBMB Imaging Platform showing that…
Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion
Researchers of the IBMB have investigated for the first time the genomic distribution of five histone H1 variants in a human cancer cell line and the consequences of H1 depletion on the 3D conformation of the genome, which will help to better understand the function of this structural component of chromatin.
Abstract
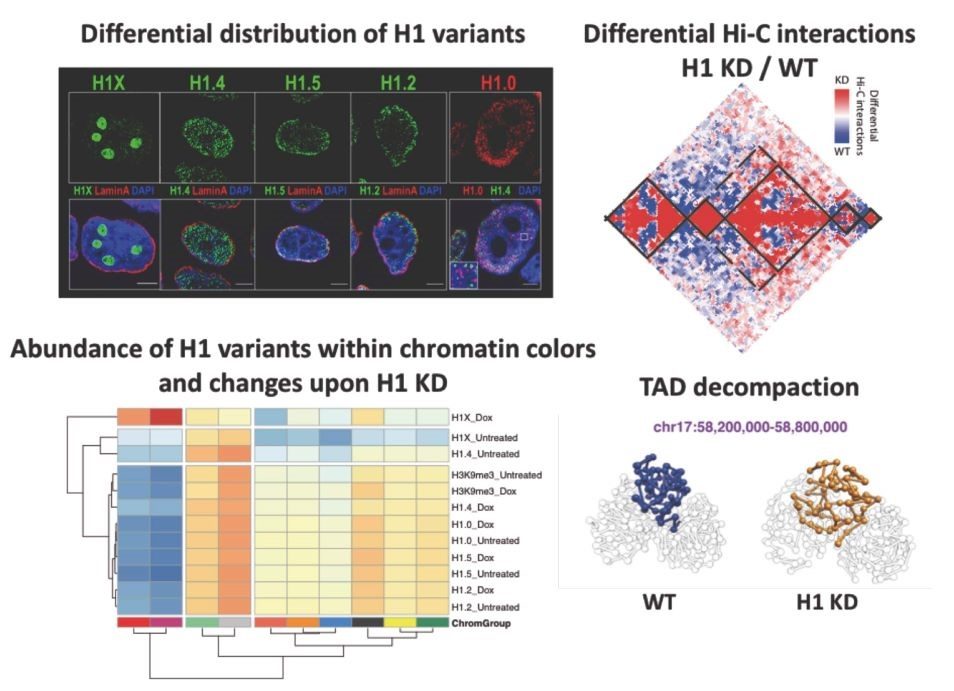
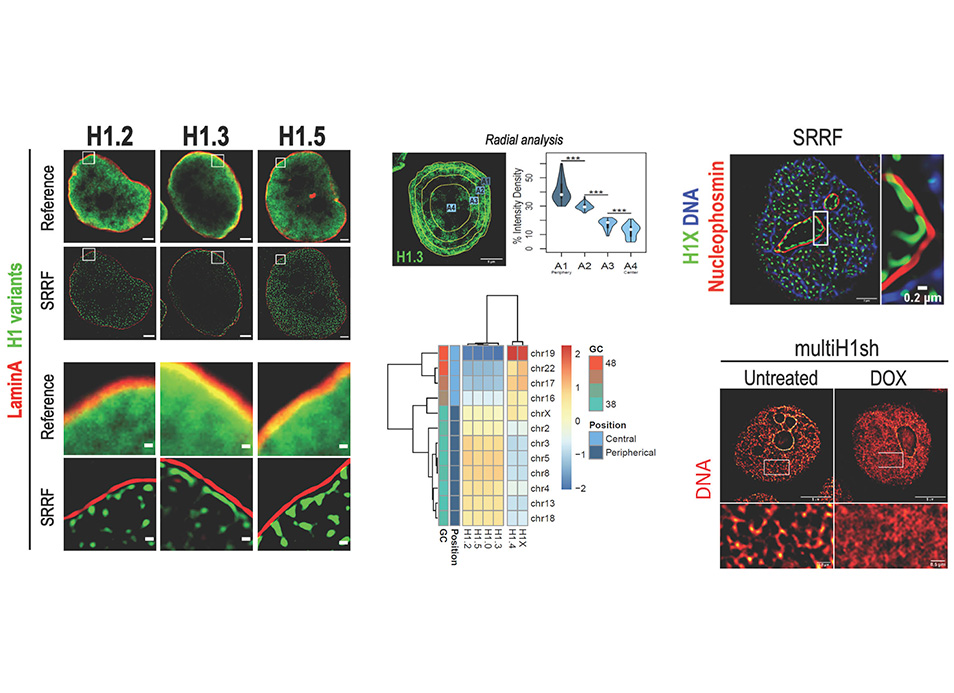
Up to seven members of the histone H1 family may contribute to chromatin compaction and its regulation in human somatic cells. In breast cancer cells, knock-down of multiple H1 variants deregulates many genes, promotes the appearance of genome-wide accessibility sites and triggers an interferon response via activation of heterochromatic repeats. However, how these changes in the expression profile relate to the re-distribution of H1 variants as well as to genome conformational changes have not been yet studied. Here, we combined ChIP-seq of five endogenous H1 variants with Chromosome Conformation Capture analysis in wild-type and H1.2/H1.4 knock-down T47D cells. The results indicate that H1 variants coexist in the genome in two large groups depending on the local GC content and that their distribution is robust with respect to H1 depletion. Despite the small changes in H1 variants distribution, knock-down of H1 translated into more isolated but de-compacted chromatin structures at the scale of topologically associating domains (TADs). Such changes in TAD structure correlated with a coordinated gene expression response of their resident genes. This is the first report describing simultaneous profiling of five endogenous H1 variants and giving functional evidence of genome topology alterations upon H1 depletion in human cancer cells.
Reference:
Serna-Pujol N*, Salinas-Pena M*, Mugianesi F*, Le Dily F, Marti-Renom MA, Jordan A (2022) Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion. Nucleic Acids Research 05 April 2022. https://doi.org/10.1093/nar/gkac226