Lab presentation
Structural Systems Biology
The cellular interior is not a mere Brownian soup of molecules; it is rather an exquisitely structured entity organized into hierarchical levels. Knowing how proteins assemble into different layers of organization, is thus fundamental to understanding the functioning of the cell.
We recently demonstrated that proteins are a few mutations, sometimes even one mutation away, to form infinite polymers –aka supramolecular assemblies– (Garcia-Seisdedos, et al. Nature 2017, Empereur-Mot, Garcia-Seisdedos, et al Sci Data 2019, Garcia-Seisdedos, et al PNAS 2022). They are amorphous or ordered structures resulting from the self-assembly of folded proteins.
In recent years a growing number of studies have identified proteins that naturally (often in a condition-dependent manner, e.g. membrane-less organelles), or as a result of mutation, form supramolecular assemblies, suggesting important implications in cellular adaptation and disease.
Although it is a widespread phenomenon that is shifting the way we see the proteome organization, supramolecular assembly remains poorly understood. Its characterization will bring about important advances with implications in evolution, disease, and protein design.
The research of the lab is at the interface of Structural, Cell, and Systems Biology and aims to understand the process by which proteins form supramolecular assemblies in the cell, as well as its role in cellular organization, adaptation, and disease. To address these aims, we combine the power of yeast genetics and high-content microscopy with biophysical and structural techniques.
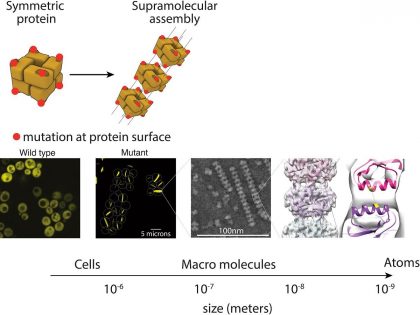
Figure adapted from Garcia-Seisdedos H, et al. Nature 2017
Projects
1 Unveiling how the spatial distribution of proteins in the cell is encoded by their sequence.
Understanding the molecular consequences of mutations in proteins is essential to map genotypes to phenotypes and interpreting the increasing wealth of genomic data. While mutations are known to disrupt protein structure and function, their potential to create new structures and localization phenotypes remains poorly understood.
Using deep mutational scanning, high-throughput microscopy, along with a variety of cellular and biophysical techniques, we address how new protein localizations and self-assembly modes are encoded in the protein sequence.
2 How does supramolecular assembly impact the cell?
To elucidate the implications of agglomeration in evolution and disease, a crucial question is how supramolecular assemblies impact the cell. Protein aggregation is paramount for cell homeostasis. Here we study whether supramolecular assemblies exhibit a similar deleterious effect or rather neutral.
We perform fitness, microscopy co-localization experiments of the assemblies with the cellular machinery (chaperones, proteasome, intracellular membrane system, etc), and evaluate whether specific knockout backgrounds sensitize cells to the formation of such assemblies. Through these experiments, we explore interaction networks connecting supramolecular assembly to the cell protein machinery.
3 Supramolecular assembly in evolution
In recent years, a growing number of studies have identified proteins that form supramolecular assemblies in response to a variety of environmental stresses such as starvation, heat shock or DNA damage. Are those assemblies playing an active role in managing cellular stress or they are rather a side effect of a cellular response to stress?
Lab people

Héctor García-Seisdedos
Hector graduated in Biology and Biochemistry from the University of Salamanca in 2007. He then moved to the Physical-Chemistry Department of the University of Granada to carry out his Ph.D. under the supervision of Beatriz Ibarra-Molero and Jose Manuel Sanchez-Ruiz. In his doctoral work, he combined experimental and bioinformatics to design tailor-made enzymes.
After earning his Ph.D. in 2012, Hector joined Emmanuel Levy’s Lab as a postdoc at the Weizmann Institute of Science in 2013, where he obtained a Koshland postdoctoral fellowship. He focused on studying how proteins self-assemble into high-order structures from a physical and cellular perspective.
In 2021, he obtained a Ramon y Cajal grant and he joined the Structural Biology Department of the IBMB in 2022 to start his group on protein assembly and evolution.
LinkedIn | ResearchGate | Twitter | CV

Meta Heidenreich
Past Students
Selected publications
*Denotes corresponding authorship
#Denotes equal contribution.
Romero-Romero ML & Garcia-Seisdedos H*. Agglomeration: When folded proteins clump together. Biophysical Reviews. 15:1987-2003. https://doi.org/10.1007/s12551-023-01172-4 (2023)
Romero-Romero ML, Garcia-Seisdedos H, Ibarra-Molero B. Active site center redesign increases protein stability preserving catalysis in thioredoxin. Protein Science 31 (9):e4447. https://doi.org/10.1002/pro.4417 (2022)
Garcia-Seisdedos H#*, Levin T#, Shapira G, Freud S, Levy ED. Mutant libraries reveal negative design shielding proteins from supramolecular self-assembly and re-localization in cells. PNAS 119 (5) e2101117119. https://doi.org/10.1073/pnas.2101117119 (2022)
Garcia-Seisdedos H, Heidenreich M, Levy ED. Not going with the flow: How cells adapt internal physics. Cell 6:183, 1462-1463. https://doi.org/10.1016/j.cell.2020.11.021 (2020)
Empereur-Mot C, Garcia-Seisdedos H, Elad N, Dei S, Levy ED. Geometric description of self-interaction potential in protein structures. Sci. Data 6:64. https://doi.org/10.1038/s41597-019-0058-x (2019)
Garcia-Seisdedos H, Villegas J, Levy ED. Infinite assembly of folded proteins in evolution, disease, and engineering. Angewandte Chemie Int Ed 58, 5514-5531. https://doi.org/10.1002/anie.201806092 (2018)
Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED. Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244-247. https://doi.org/10.1038/nature23320 (2017)
+2 F1000 recommendations
Garcia-Seisdedos H, Steinberg A, Levy ED. Symmetry breaking in homo-oligomers: The curious case of mega-hemocyanin. Structure 23:1, 3-5. https://doi.org/10.1016/j.str.2014.12.006 (2015)
Garcia-Seisdedos H, Ibarra-Molero B & Sanchez-Ruiz JM. Probing the mutational interplay between primary and promiscuous protein functions: a computational-experimental approach, PLoS Comput Biol 8, e1002558. https://doi.org/10.1371/journal.pcbi.1002558 (2012)
Garcia-Seisdedos H, Ibarra-Molero B & Sanchez-Ruiz JM. How many ionizable groups can sit on a protein hydrophobic core? Proteins 80, 1-7. https://doi.org/10.1002/prot.23166 (2012)
All publications
*Denotes corresponding authorship
#Denotes equal contribution.
Romero-Romero ML & Garcia-Seisdedos H*. Agglomeration: When folded proteins clump together. Biophysical Reviews. 15:1987-2003. https://doi.org/10.1007/s12551-023-01172-4 (2023)
Romero-Romero ML, Garcia-Seisdedos H, Ibarra-Molero B. Active site center redesign increases protein stability preserving catalysis in thioredoxin. Protein Science 31 (9):e4447. https://doi.org/10.1002/pro.4417 (2022)
Garcia-Seisdedos H#*, Levin T#, Shapira G, Freud S, Levy ED. Mutant libraries reveal negative design shielding proteins from supramolecular self-assembly and re-localization in cells. PNAS 119 (5) e2101117119. https://doi.org/10.1073/pnas.2101117119 (2022)
Garcia-Seisdedos H, Heidenreich M, Levy ED. Not going with the flow: How cells adapt internal physics. Cell 6:183, 1462-1463. https://doi.org/10.1016/j.cell.2020.11.021 (2020)
Empereur-Mot C, Garcia-Seisdedos H, Elad N, Dei S, Levy ED. Geometric description of self-interaction potential in protein structures. Sci. Data 6:64. https://doi.org/10.1038/s41597-019-0058-x (2019)
Garcia-Seisdedos H, Villegas J, Levy ED. Infinite assembly of folded proteins in evolution, disease, and engineering. Angewandte Chemie Int Ed 58, 5514-5531. https://doi.org/10.1002/anie.201806092 (2018)
Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED. Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244-247. https://doi.org/10.1038/nature23320 (2017)
+2 F1000 recommendations
Garcia-Seisdedos H, Steinberg A, Levy ED. Symmetry breaking in homo-oligomers: The curious case of mega-hemocyanin. Structure 23:1, 3-5. https://doi.org/10.1016/j.str.2014.12.006 (2015)
Garcia-Seisdedos H, Ibarra-Molero B & Sanchez-Ruiz JM. Probing the mutational interplay between primary and promiscuous protein functions: a computational-experimental approach, PLoS Comput Biol 8, e1002558. https://doi.org/10.1371/journal.pcbi.1002558 (2012)
Garcia-Seisdedos H, Ibarra-Molero B & Sanchez-Ruiz JM. How many ionizable groups can sit on a protein hydrophobic core? Proteins 80, 1-7. https://doi.org/10.1002/prot.23166 (2012)
Project funding
Ongoing Projects
Tittle: “Infinite assembly of folded proteins”
Dates: 2022-2025
Role: PI
Proyecto PID2021-127150NA-I00 financiado por:

Ramon y Cajal research grant, Ed. 2020
Dates: 2022-2027
Role: PI
Ayuda RYC2020-030700-I financiada por:

Vacancies/Jobs
We are currently seeking motivated and creative researchers at all career stages to join the lab. Please, contact Hector Garcia (hgsbmc@ibmb.csic.es) for more details.
Lab corner
Project gallery
No albums or photos uploaded yet.
