Lab presentation
Our research focuses on the structure-function relationships of proteins and macromolecular complexes directly implicated in pathological processes. In particular, virus particles, viral proteins and protein-complexes involved in RNA replication as well as other large macromolecular assemblies. The information obtained would be instrumental not only for improving the knowledge about the functioning of these biological systems but also in the development of novel therapeutic strategies. We use a number of molecular and structural biology techniques. In particular, X-ray crystallography and combinations of X-ray crystallography and Cryo-electron microscopy.
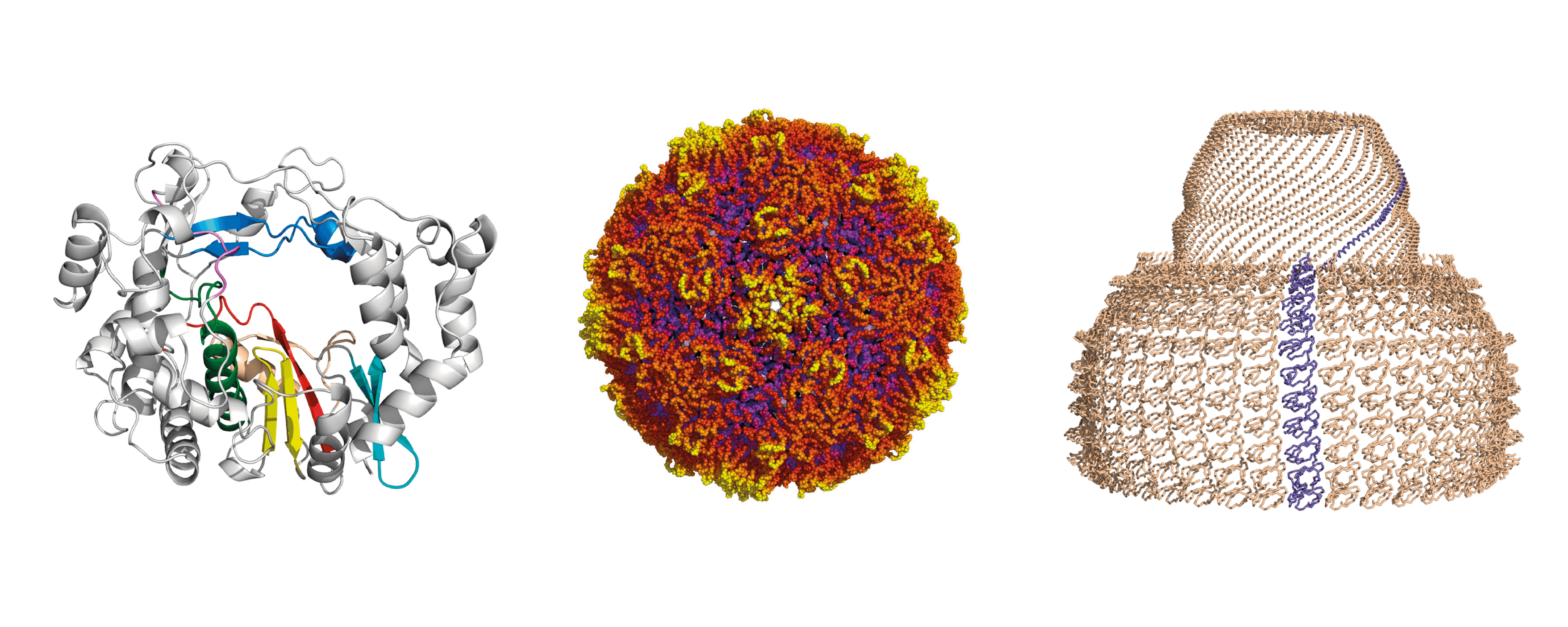
Projects
- Structural and functional studies of viral RNA-dependent RNA polymerases and replication complexes
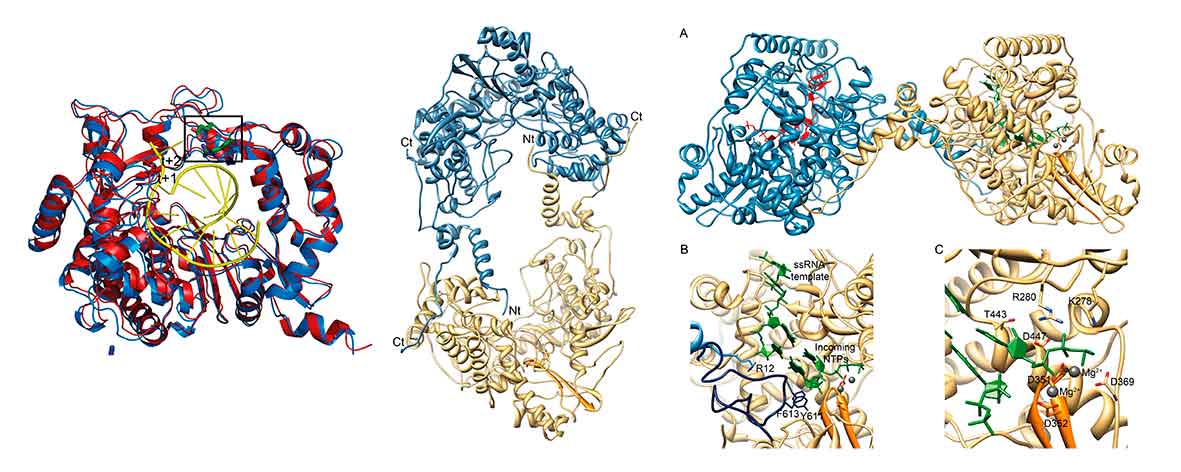
Most emerging and re-emerging human and animal viral diseases are associated with RNA viruses. One way to limit the impact of these pathogens is to prevent their replication and a thorough understanding of their replication and transcription machineries is therefore essential. Despite this group of viruses use different strategies for genome replication and transcription all RNA virus families depend on the activity of a virally encoded RNA-dependent RNA polymerase (RdRP). The structural information obtained for different RdRP and RdRP-RNA complexes contributed decisively to our knowledge about the functioning these enzymes. Despite the many clues provided by these structures there are still several gaps in our knowledge about the regulation of the RdRP activity and fidelity in different virus families.
We are combining structural data and functional studies to elucidate how RdRPs interact with RNA promotors, other viral proteins and cellular factors, to regulate replication and transcription activities in diferent viruses, including picornaviruses, flaviviruses, coronaviruses and filoviruses. This information will contribute decisively to our knowledge about the mechanisms of action of these enzymes, paving the way for the development of new therapeutic strategies against these pathogens.
- Protein-protein interactions in viral replication and pathogenesis
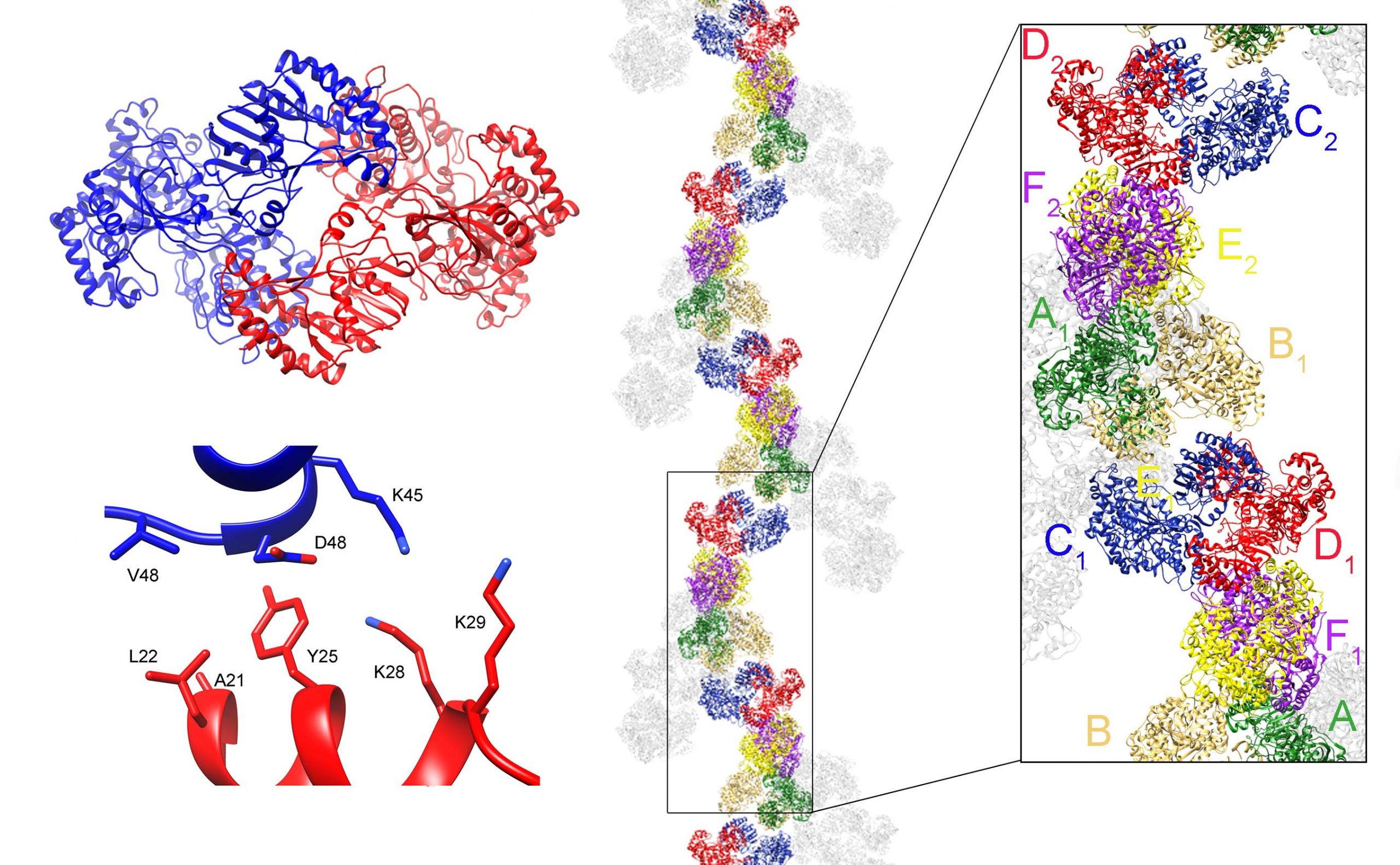
RNA viruses usually associate the RdRPs to other viral or host proteins as scaffold to form the replicative complexes in specific intracellular locations like the membranous compartments or inside capsids. The RdRP self-interactions to form dimers and higher order oligomers also play an important role in viral replication, contributing to the fine tuning of the polymerase activity, ensuring the proper levels of RNA production.
We have previous characterized the structure and quaternary arrangement of the Methyltransferase-RdRP polypeptide NS5 from Zika virus (ZIKV), showing that this protein is able to form dimers and higher-order oligomers that regulate its enzymatic functions, as well as its association with other viral or host factors during infection. More recently we have shown that the multimeric arrangement of ZIKV-NS5 is required for nuclear exit, localization and interaction with primary cilium base proteins, and for promoting ciliopathy and premature neurogenesis. We have also showed that ZIKV-NS5 interacted with and depleted a number of cilia/the centrosome proteins (including, Rootletin, BART1, CEP164) and we aim to structurally and functionally characterize these interactions as new potential targets for therapeutic intervention.
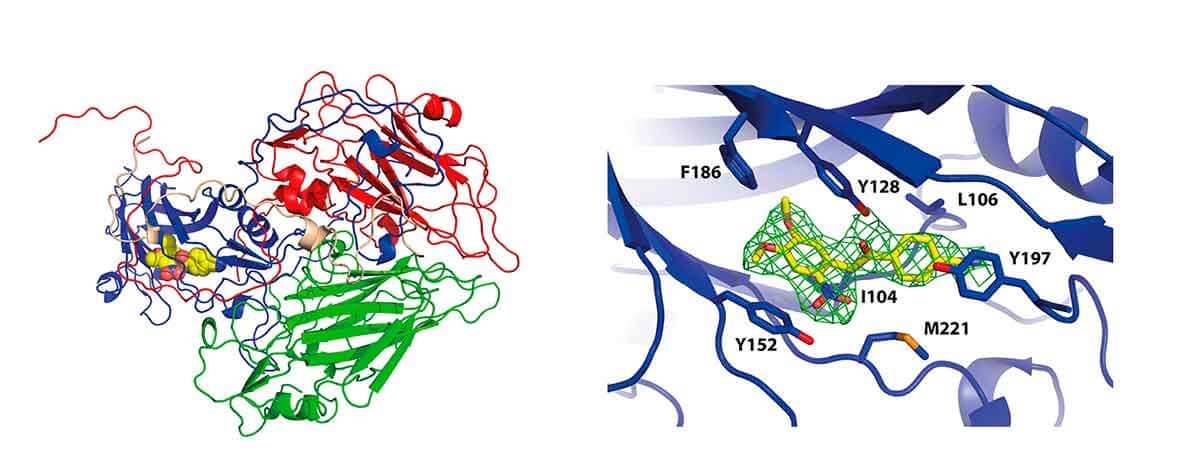
- Structure and dynamics of viral capsids
We have previously characterized the early steps of human rhinovirus infection, including receptor binding by solving the first high resolution X-ray structures of a human minor group rhinovirus (RV1A) bound to its cellular receptor protein. We have also determined the structural rearrangements occurring in the RV1A capsid trapped in the process of RNA uncoating. We are currently studying the structural dynamics of other RV capsids, including the effect of a number of capsid binders as antiviral candidates, preventing RNA uncoating.
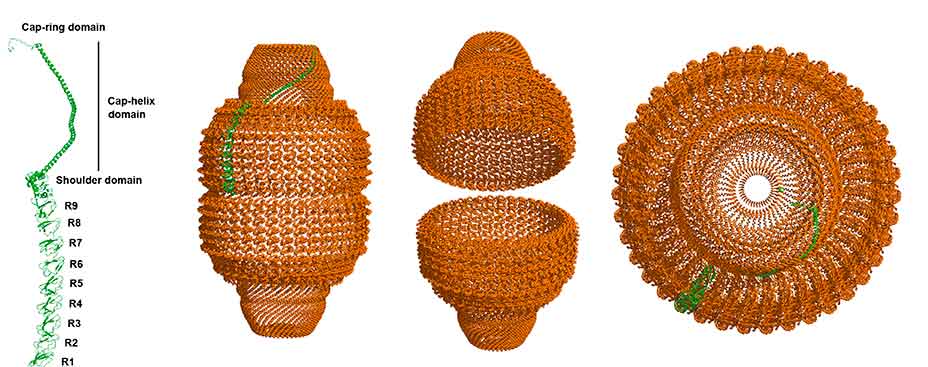
- Structural dynamics of the vault shell
Vaults are ubiquitous ribonucleoprotein particles involved in a diversity of cellular processes, with promising applications as nanodevices for delivery of multiple cargos. The vault shell is assembled by the symmetrical association of multiple copies of the major vault protein that, initially, generates half vaults. The pairwise, anti-parallel association of two half vaults produces the whole particle. Using a combination of biophysical and structural techniques with the production of recombinant mutant vaults, we aim to decipher the principles of vault dynamics and opening.
Lab people

Nuria Verdaguer Massana
Núria Verdaguer is the Scientific Director of the of the Maria de Maeztu Structural Biology Unit at the Institut de Biologia Molecular de Barcelona (IBMB-CSIC).
She obtained her degree in Biological Sciences from the University of Barcelona (UB) in 1986 and her Ph.D. in 1991 from the Polytechnic University of Catalonia (UPC). In 2002 she started a structural virology research group at the IBMB-CSIC and promoted to full Professor in 2009.
Núria Verdaguer was elected member of the European Molecular Biology Organization (EMBO) in 2008. She has been director of the Department of Structural Biology from October 2010 to September 2012 and Deputy Director of the IBMB from July 2014 to June 2016.

Diego Sebastian Ferrero

Cristina Ferrer Orta

María González

Sergi Vazquez
Past students
Omar Tomás
Victor Manuel Ruiz Arroyo
Selected publications
Structural insights on the nucleoprotein C-terminal domain of Měnglà virus
Ferrero DS, Tomás Gilabert O, Verdaguer N.
Microbiol Spectr 2023 11:e02373-23. doi: 10.1128/spectrum.02373-23.
Symmetry disruption commits vault particles to disassembly.
Guerra P, González-Alamos M, Llauró A, Casañas A, Querol-Audí J, de Pablo PJ, Verdaguer N.
Sci Adv. 2022 Feb 11;8(6):eabj7795. doi: 10.1126/sciadv.abj7795.
Multimerization of Zika Virus-NS5 Causes Ciliopathy and Forces Premature Neurogenesis.
Saade M, Ferrero DS, Blanco-Ameijeiras J, Gonzalez-Gobartt E, Flores-Mendez M, Ruiz-Arroyo VM, Martínez-Sáez E, Ramón Y Cajal S, Akizu N, Verdaguer N, Martí E. Cell Stem Cell. 2020 Dec 3;27(6):920-936.e8. doi: 10.1016/j.stem.2020.10.002.
Highlighted in Nature Rev Microbiol https://www.nature.com/articles/s41579-020-00481-9
Supramolecular arrangement of the full-length Zika virus NS5.
Ferrero DS, Ruiz-Arroyo VM, Soler N, Usón I, Guarné A, Verdaguer N.
PLoS Pathog. 2019 Apr 5;15(4):e1007656.
Structural characterization of the Rabphilin-3A-SNAP25 interaction.
Ferrer-Orta C, Pérez-Sánchez MD, Coronado-Parra T, Silva C, López-Martínez D, Baltanás-Copado J, Gómez-Fernández JC, Corbalán-García S, Verdaguer N.
Proc Natl Acad Sci U S A. 2017 Jul 3;114(27):E5343-E5351.
Ferrero DS, Buxaderas M, Rodríguez JF, Verdaguer N.
PLoS Pathog. 2015 Dec 1;11(12):e1005265.
Guillén J, Ferrer-Orta C, Buxaderas M, Pérez-Sánchez D, Guerrero-Valero M, Luengo-Gil G, Pous J, Guerra P, Gómez-Fernández JC, Verdaguer N*, Corbalán-García S* (*corresponding Authors).
Proc Natl Acad Sci USA. 2013 Dec 17;110(51):20503-8.
Pickl-Herk A, Luque D, Vives-Adrián L, Querol-Audí J, Garriga D, Trus BL, Verdaguer N*, Blaas D*, Castón JR* (*corresponding Authors).
Proc Natl Acad Sci USA.2013 Dec 10;110(50):20063-8.
Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid
Garriga D, Pickl-Herk A, Luque D, Wruss J, Castón JR, Blaas D, Verdaguer N.
PLoS Pathog. 2012 Jan;8(1):e1002473.
The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Querol-Audí J, Casañas A, Usón I, Luque D, Castón JR, Fita I, Verdaguer N.
EMBO J. 2009 Nov 4;28(21):3450-7.
Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2
Guerrero-Valero M, Ferrer-Orta C, Querol-Audí J, Marin-Vicente C, Fita I, Gómez-Fernández JC, Verdaguer N*, Corbalán-García S*(*corresponding authors).
Proc Natl Acad Sci USA. 2009 Apr 21;106(16):6603-7.
Activation mechanism of a noncanonical RNA-dependent RNA polymerase
Garriga D, Navarro A, Querol-Audí J, Abaitua F, Rodríguez JF, Verdaguer N.
Proc Natl Acad Sci. USA. 2007;104:20540-5.
Sequential structures provide insights into the fidelity of RNA replication
Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N.
Proc Natl Acad Sci. USA. 2007;104:9463-8
The structure of a protein primer-polymerase complex in the initiation of genome replication
Ferrer-Orta C, Arias A, Agudo R, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N.
EMBO J. 2006 Feb 22;25(4):880-8.
Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D.
Nat Struct Mol Biol. 2004; 11:429-34.
All publications
Structural insights on the nucleoprotein C-terminal domain of Měnglà virus
Ferrero DS, Tomás Gilabert O, Verdaguer N.
Microbiol Spectr 2023 11:e02373-23. doi: 10.1128/spectrum.02373-23.
Symmetry disruption commits vault particles to disassembly.
Guerra P, González-Alamos M, Llauró A, Casañas A, Querol-Audí J, de Pablo PJ, Verdaguer N. Sci Adv. 2022 Feb 11;8(6):eabj7795. doi: 10.1126/sciadv.abj7795.
Special Issue: “Viral Replication Complexes”.
Verdaguer N, Ferrero DS.
Viruses. 2021 Sep 23;13(10):1902. doi: 10.3390/v13101902.
Snapshots of a Non-Canonical RdRP in Action.
Ferrero DS, Falqui M, Verdaguer N.
Viruses. 2021 Jun 28;13(7):1260. doi: 10.3390/v13071260.
Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
Ferrero DS, Busnadiego I, Garriga D, Guerra P, Martín MT, Kremer L, Usón I, Rodriguez JF, Verdaguer N.
J Virol. 2020 Nov 25;95(4):e02166-20. doi: 10.1128/JVI.02166-20.
Multimerization of Zika Virus-NS5 Causes Ciliopathy and Forces Premature Neurogenesis.
Saade M, Ferrero DS, Blanco-Ameijeiras J, Gonzalez-Gobartt E, Flores-Mendez M, Ruiz-Arroyo VM, Martínez-Sáez E, Ramón Y Cajal S, Akizu N, Verdaguer N, Martí E.
Cell Stem Cell. 2020 Dec 3;27(6):920-936.e8. doi:10.1016/j.stem.2020.10.002.
Amino Acid Substitutions Associated with Treatment Failure for Hepatitis C Virus Infection.
Soria ME, García-Crespo C, Martínez-González B, Vázquez-Sirvent L, Lobo-Vega R, de Ávila AI, Gallego I, Chen Q, García-Cehic D, Llorens-Revull M, Briones C, Gómez J, Ferrer-Orta C, Verdaguer N, Gregori J, Rodríguez-Frías F, Buti M, Esteban JI, Domingo E, Quer J, Perales C.
J Clin Microbiol. 2020 Nov 18;58(12):e01985-20. doi: 10.1128/JCM.01985-20.
Wald J, Pasin M, Richter M, Walther C, Mathai N, Kirchmair J, Makarov VA, Goessweiner-Mohr N, Marlovits TC, Zanella I, Real-Hohn A, Verdaguer N, Blaas D, Schmidtke M.
Proc Natl Acad Sci U S A. 2019 Sep 17;116(38):19109-19115. doi: 10.1073/pnas.1904732116.
de Castro S, Ferrer-Orta C, Mills A, Fernández-Cureses G, Gago F, Verdaguer N, Camarasa MJ.
Molecules. 2019 Jun 26;24(13). pii: E2360. doi: 10.3390/molecules24132360.
Supramolecular arrangement of the full-length Zika virus NS5.
Ferrero DS, Ruiz-Arroyo VM, Soler N, Usón I, Guarné A, Verdaguer N.
PLoS Pathog. 2019 Apr 5;15(4):e1007656.
de la Higuera I, Ferrer-Orta C, Moreno E, de Ávila AI, Soria ME, Singh K, Caridi F, Sobrino F, Sarafianos SG, Perales C, Verdaguer N, Domingo E.
J Virol. 2018 Sep 26;92(20). pii: e01119-18. doi: 10.1128/JVI.01119-18.
Viral RNA-Dependent RNA Polymerases: A Structural Overview
Ferrero D, Ferrer-Orta C, Verdaguer N.
Subcell Biochem. 2018;88:39-71.
Structural characterization of the Rabphilin-3A-SNAP25 interaction.
Ferrer-Orta C, Pérez-Sánchez MD, Coronado-Parra T, Silva C, López-Martínez D, Baltanás-Copado J, Gómez-Fernández JC, Corbalán-García S, Verdaguer N.
Proc Natl Acad Sci U S A. 2017 Jul 3;114(27):E5343-E5351.
Miras M, Truniger V, Silva C, Verdaguer N, Aranda MA, Querol-Audí J.
Plant Physiol. 2017 Jul;174(3):1476-1491
Guerra P, Valbuena A, Querol-Audí J, Silva C, Castellanos M, Rodríguez-Huete A, Garriga D, Mateu MG, Verdaguer N.
Sci Rep. 2017 Jun 22;7(1):4101.
Molecular and Functional Bases of Selection against a Mutation Bias in an RNA Virus.
de la Higuera I, Ferrer-Orta C, de Ávila AI, Perales C, Sierra M, Singh K, Sarafianos SG, Dehouck Y, Bastolla U, Verdaguer N, Domingo E.
Genome Biol Evol. 2017 May 1;9(5):1212-1228.
Llauró A, Guerra P, Kant R, Bothner B, Verdaguer N.
Sci Rep. 2016 Oct 14:6:34143.
Herod MR, Ferrer-Orta C, Loundras EA, Ward JC, Verdaguer N, Rowlands DJ, Stonehouse NJ.
J Virol. Jul 11;90(15):6864-83. (2016)
Ferrero DS, Buxaderas M, Rodríguez JF, Verdaguer N.
PLoS Pathog. Dec 1;11(12):e1005265. (2015)
Infectious Bursal Disease Virus VP3 Upregulates VP1-Mediated RNA-Dependent RNA Replication.
Ferrero D, Garriga D, Navarro A, Rodríguez JF, Verdaguer N.
J Virol. Nov;89(21):11165-8. (2015)
RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms.
Ferrer-Orta C, Ferrero D, Verdaguer N.
Viruses. Aug 6;7(8):4438-60. (2015)
Ferrer-Orta C, de la Higuera I, Caridi F, Sánchez-Aparicio MT, Moreno E, Perales C, Singh K, Sarafianos SG, Sobrino F, Domingo E, Verdaguer N.
J Virol. Apr 22. pii: JVI.03283-14. (2015)
Van der Linden L, Vives-Adrián L, Selisko B, Ferrer-Orta C, Liu X, Lanke K, Ulferts R, De Palma AM, Tanchis F, Goris N, Lefebvre D, De Clercq K, Leyssen P, Lacroix C, Pürstinger G, Coutard B, Canard B, Boehr DD, Arnold JJ, Cameron CE, Verdaguer N, Neyts J, van Kuppeveld FJ.
PLoS Pathog. Mar 23;11(3):e1004733. (2015)
Structural basis for host membrane remodeling induced by protein 2B of hepatitis A virus.
Vives-Adrián L, Garriga D, Buxaderas M, Fraga J, Pereira PJ, Macedo-Ribeiro S, Verdaguer N.
J Virol. Apr;89(7):3648-58. (2015)
A novel benzonitrile analogue inhibits rhinovirus replication.
Verdaguer N, Ferrero D, Murthy MR. Viruses and viral proteins.
IUCrJ. Oct 14;1(Pt 6):492-504. (2014)
A novel benzonitrile analogue inhibits rhinovirus replication
Lacroix C, Querol-Audí J, Roche M, Franco D, Froeyen M, Guerra P, Terme T,Vanelle P, Verdaguer N, Neyts J, Leyssen P.
J Antimicrob Chemother. Oct;69(10):2723-32. (2014)
Luque D, Gómez-Blanco J, Garriga D, Brilot AF, González JM, Havens WM, Carrascosa JL, Trus BL, Verdaguer N, Ghabrial SA, Castón JR.
Proc Natl Acad Sci USA. May 27;111(21):7641-6. (2014)
Vives-Adrian L, Lujan C, Oliva B, van der Linden L, Selisko B, Coutard B, Canard B, van Kuppeveld FJ, Ferrer-Orta C, Verdaguer N.
J Virol. May;88(10):5595-607. (2014)
Mechanical stability and reversible fracture of vault particles.
Llauró A, Guerra P, Irigoyen N, Rodríguez JF, Verdaguer N, de Pablo PJ.
Biophys J. Feb 4;106(3):687-95. (2014)
Guillén J, Ferrer-Orta C, Buxaderas M, Pérez-Sánchez D, Guerrero-Valero M, Luengo-Gil G, Pous J, Guerra P, Gómez-Fernández JC, Verdaguer N*, Corbalán-García S* (*corresponding Authors).
Proc Natl Acad Sci USA. Dec17;110(51):20503-8. (2013)
Pickl-Herk A, Luque D, Vives-Adrián L, Querol-Audí J, Garriga D, Trus BL, Verdaguer N*, Blaas D*, Castón JR* (*corresponding Authors).
Proc Natl Acad Sci USA. Dec 10;110(50):20063-8. (2013)
X-ray crystallography of viruses.
Verdaguer N, Garriga D, Fita I.
Subcell Biochem. 68:117-44. (2013)
Casañas A, Querol-Audí J, Guerra P, Pous J, Tanaka H, Tsukihara T, Verdaguer N*, Fita I* (*corresponding authors).
Acta Crystallogr D BiolCrystallogr. Jun;69(Pt6):1054-61. (2013)
Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity.
Garriga D, Ferrer-Orta C, Querol-Audí J, Oliva B, Verdaguer N.
J Mol Biol. Jul 10;425(13):2279-87. (2013)
Ferrero D, Buxaderas M, Rodriguez JF, Verdaguer N.
Acta Crystallogr Sect F Struct Biol Cryst Commun. Oct 1;68(Pt 10):1263-6. (2012)
Conformational changes in motif D of RdRPs as fidelity determinant.
Verdaguer N, Ferrer-Orta C.
Structure. Sep 5;20(9):1448-50. (2012)
Vault particles: a new generation of delivery nanodevices.
Casañas A, Guerra P, Fita I, Verdaguer N.
Curr Opin Biotechnol. Dec;23(6):972-7 (2012)
Luque D, González JM, Gómez-Blanco J, Marabini R, Chichón J, Mena I, Angulo I, Carrascosa JL, Verdaguer N, Trus BL, Bárcena J, Castón JR.
J Virol. Jun;86(12):6470-80. (2012)
Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid.
Garriga D, Pickl-Herk A, Luque D, Wruss J, Castón JR, Blaas D, Verdaguer N.
PLoS Pathog. Jan;8(1):e1002473. (2012)
Garriga D, Vives-Adrián L, Buxaderas M, Ferreira-da-Silva F, Almeida B, Macedo-Ribeiro S, Pereira PJ, Verdaguer N.
Acta Crystallogr Sect F Struct Biol Cryst Commun. Oct 1;67(Pt 10):1224-7. (2011)
Agudo R, Ferrer-Orta C, Arias A, de la Higuera I, Perales C, Pérez-Luque R, Verdaguer N, Domingo E.
PLoS Pathog. Aug 26;6(8):e1001072. (2010)
Luque D, González JM, Garriga D, Ghabrial SA, Havens WM, Trus B, Verdaguer N, Carrascosa JL, Castón JR.
J Virol. Jul;84(14):7256-66. (2010)
Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin.
Ferrer-Orta C, Sierra M, Agudo R, de la Higuera I, Arias A, Pérez-Luque R, Escarmís C, Domingo E, Verdaguer N.
J Virol. Jun;84(12):6188-99. (2010)
Ferrer-Orta C, Agudo R, Domingo E, Verdaguer N.
Curr Opin Struct Biol. Dec;19(6):752-8. (2009)
The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP.
Querol-Audí J, Casañas A, Usón I, Luque D, Castón JR, Fita I, Verdaguer N.
EMBO J. Nov 4;28(21):3450-7. (2009)
Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2.
Guerrero-Valero M, Ferrer-Orta C, Querol-Audí J, Marin-Vicente C, Fita I, Gómez-Fernández JC, Verdaguer N*, Corbalán-García S*(*corresponding authors).
Proc Natl Acad Sci USA. Apr 21;106(16):6603-7. (2009)
Autoproteolytic activity derived from the infectious bursal disease virus capsid protein.
Irigoyen N, Garriga D, Navarro A, Verdaguer N, Rodríguez JF, Castón JR.
J Biol Chem. Mar 20;284(12):8064-72. (2009)
Querol-Audí J, Konecsni T, Pous J, Carugo O, Fita I, Verdaguer N, Blaas D.
FEBS Lett. Jan 5;583(1):235-40. (2009)
Structural insights into the multifunctional protein VP3 of birnaviruses.
Casañas A, Navarro A, Ferrer-Orta C, González D, Rodríguez JF, Verdaguer N.
Structure. 16:29-37. (2008)
Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Garriga D, Navarro A, Querol-Audí J, Abaitua F, Rodríguez JF, Verdaguer N.
Proc Natl Acad Sci. USA. 104:20540-5. (2007)
Sequential structures provide insights into the fidelity of RNA replication.
Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N.
Proc Natl Acad Sci. USA. 104:9463-8 (2007)
Luque D, Saugar I, Rodriguez JF, Verdaguer N, Garriga D, Martin CS, Velazquez-Muriel JA, Trus BL, Carrascosa JL, Caston JR.
J Virol. 81:6869-78. (2007)
Garriga D, Querol-Audi J, Abaitua F, Saugar I, Pous J, Verdaguer N, Caston JR, Rodriguez JF.
J Virol. 80:6895-905. (2006)
The structure of a protein primer-polymerase complex in the initiation of genome replication.
Ferrer-Orta C, Arias A, Agudo R, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N.
EMBO J. Feb 22;25(4):880-8. (2006)
A comparison of viral RNA-dependent RNA polymerases.
Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N.
Curr Opin Struct Biol. 16:27-34. (2006)
Arias A, Agudo R, Ferrer-Orta C, Perez-Luque R, Airaksinen A, Brocchi E, Domingo E, Verdaguer N, Escarmis C.
J Mol Biol. 353:1021-32. (2005)
Preliminary analysis of two and three dimensional crystals of vault ribonucleoprotein particles.
Querol-Audi J, Perez-Luque R, Fita I, Lopez-Iglesias C, Caston JR, Carrascosa JL, Verdaguer N.
J Struct Biol. 151:111-5. (2005)
Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N.
J Biol Chem. 279:47212-21. (2004)
Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D.
Nat Struct Mol Biol. 11:429-34. (2004)
Barcena J, Verdaguer N, Roca R, Morales M, Angulo I, Risco C, Carrascosa JL, Torres JM, Caston JR.
Virology. 322:118-34. (2004)
Crystallization and preliminary X-ray analysis of the glycogen synthase from Pyrococcus abyssi.
Horcajada C, Cid E, Guinovart JJ, Verdaguer N, Ferrer JC.
Acta Crystallogr D Biol Crystallogr. 59:2322-4. (2003)
Evolution of cell recognition by viruses: a source of biological novelty with medical implications.
Baranowski E, Ruiz-Jarabo CM, Pariente N, Verdaguer N, Domingo E.
Adv Virus Res. 62:19-111. (2003)
Verdaguer N, Jimenez-Clavero MA, Fita I, Ley V.
J Virol 77:9780-9. (2003)
Retinoic acid binds to the C2-domain of protein kinase C(alpha).
Ochoa WF, Torrecillas A, Fita I, Verdaguer N, Corbalan-Garcia S, Gomez-Fernandez JC.
Biochemistry. 42:8774-9. (2003)
Crystallization and preliminary X-ray analysis of swine vesicular disease virus (SVDV).
Jimenez-Clavero MA, Ley V, Fita I, Verdaguer N.
Acta Crystallogr D Biol Crystallogr. 59:541-3. (2003)
W.F.Ochoa, S. Corbalan-Garcia, R. Eritja, J.C. Gomez-Fernandez, I. Fita & N.Verdaguer
J. Mol. Biol. 320:277-291. (2002)
P.Gomes, W. F., Ochoa, N.Verdaguer, E. Giralt and D. Andreu.
J. Pep. Res. 59: 221-231. (2003)
W.F.Ochoa, J. Garcia-Garcia, I. Fita, S. Corbalan-Garcia, N. Verdaguer, J.C. Gomez-Fernandez.
J. Mol. Biol. 311:837-849 (2001)
R. Perez-Luque, X.Carpena., W.F. Ochoa., N. Verdaguer., W. Melik-Adamyan, M. Klotz, J. Switala, I. Fita. & P.Loewen.
Acta Cryst. 2001; D57: 1184-1186. (2001)
S.G. Kalko, M. Chagoyen, N. Jimenez-Lozano, N. Verdaguer, I. Fita., J.M. Carazo.
Eur. Biophys J. 29: 457-462 (2000)
W.F.Ochoa, S. Kalko, M.G.Mateu, P.Gomes, D.Andreu, E.Domingo, I.Fita and N.Verdaguer.
J.Gen.Virol. 81: 1495-1505. (2000)
Structure of human rhinovirus serotype 2 (HRV2)
N.Verdaguer, D.Blaas, I.Fita.
J. Mol. Biol. 300:1179-1194. (2000)
N.Verdaguer, G.Schoehn, W.F.Ochoa, I.Fita, S.Brooks, A.King, E.Domingo, M.G.Mateu, D.Stuart, E.Hewat.
Virology 255:260-268. (1999)
Crystallization and preliminar x-ray analysis of human rhinovirus serotype 2 (HRV2).
N.Verdaguer, T. Marlovits, J.Bravo, D.Stuart, D.Blaas and I.Fita.
Acta Cryst D54:1459-1461. (1999)
Ca2+ bridges the membrane-binding domain C2 od Protein Kinase Ca directly tom phosphatidylserine
N.Verdaguer, S. Corbalan-Garcia, W.F. Ochoa, I. Fita, J.C. Gomez-Fernandez.
EMBO J. 18: 6329-6338. (1999)
E.Domingo, N. Verdaguer, W.F.Ochoa, C.M. Ruiz-Jarabo,N. Sevilla, E.Baranowski, D.Andreu, M.G.Mateu, I.Fita.
Virus Research 62: 169-175. (1999)
N. Verdaguer, N. Sevilla, M.L. Valero, D. Stuart, E. Brocchi, D. Andreu, E. Giralt, E. Domingo, M. Mateu and I.Fita.
J Virol 72:739-748. (1998)
Multiple Virulence Determinants of Foot-and-Mouth Disease Virus in Cell Culture
E.Baranowski, N. Sevilla, N.Verdaguer, C.M. Ruiz-Jarabo, E.Beck, E.Domingo
JVirol 72:6362-6372. (1998)
C.Escarmís, E.C. Carrillo, M.Ferrer, J.F.Garcia Arriaza, N. López, Cecilia Tami, N. Verdaguer, E.Domingo and M.T.Fanze-Fernandez.
J Virol 72:10171-10179. (1998)
E.A. Hewat, N. Verdaguer, I.Fita, W. Blakemore, S. Brookers, A. King, J. Newman, E. Domingo, M.G. Mateu, D. Stuart.
EMBO Journal 16:1492-1500. (1997)
M.A.Martinez, N.Verdaguer, M.G.Mateu, E.Domingo
Proc Natl Acad Sci USA. 94:6798-6802. (1997)
Efficient Neutralization of Foot-and-Mouth Disease Virus by Monovalent Antibody Binding.
N.Verdaguer, I.Fita, E.Domingo, M.G.Mateu.
J Virol 71: 9813-9816. (1997)
Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG).
L.M.Gonzaga, J.A. Subirana, N.Verdaguer, D. Pyshnyi, L.Campos, L. Malinina.
Journal of Biomolecular Structure and dynamics 15:151-163. (1997)
Structure and supramolecualr Packing features of the dipeptide Arg-Val acetate.
R.Recacha, N. Verdaguer and J.A. Subirana.
Int. J. pept. Prot. Res. 50:388-392. (1997)
N.Verdaguer, M.G.Mateu, J.Bravo, E.Domingo, I.Fita.
J Mol Biol, 256:364-376. (1996)
Molecular Evolution of Aphthoviruses.
- Domingo, M.G.Mateu, C. Escarmis, E.Martínez-Salas, D.Andreu, E. Giralt, N. Verdaguer, I.Fita.
Virus Genes 11:197-207. (1996)
A.Guarné, R.Kirchweger, N.Verdaguer, H-D.Liebig, D.Blaas, T.Skern, I.Fita.
Protein Science 5:1931-1933. (1996)
N.Sevilla, N.Verdaguer, E.Domingo.
Virology 225:400-405. (1996)
N. Verdaguer, M.G. Mateu, D. Andreu, E. Giralt, E. Domingo, I.Fita.
EMBO Journal 14: 1690-1696. (1995)
Crystal structure of catalase HPII from Escherichia coli.
J. Bravo, N. Verdaguer, J. Tormo, C. Betzel, J. Switala, P.C. Loewen, I.Fita.
Structure 3:491-502. (1995)
M.L. Valero, J.A. Camarero, A. Adeva, N. Verdaguer, I.Fita, M.G. Mateu, E. Domingo, E. Giralt, D. Andreu.
Biomedical Peptides, Proteins & Nucleic Acids 1:133-140. (1995)
N. Verdaguer, M.G. Mateu, J. Bravo, J. Tormo, E. Giralt, D. Andreu, E. Domingo, I.Fita.
Proteins: Structure, Function and Genetics 18:201-203. (1994)
Structure of the dipeptide L-lysine-L-leucine. Acetate, 0.5 acetic acid hemihydrate.
M. Perelló, N. Verdaguer, J. Aymami, I.Fita.
Journal of Chemical Crystallography 24:597-602. (1994)
L.Tabernero, N.Verdaguer, M.Coll,,G.A. van der Marel, J.H.van Boom, A.Rich and J.Aymamí.
Biochemistry 32:8403-8410. (1993)
Helical structure of basic proteins from spermatozoa. Comparison with model peptides.
N. Verdaguer, M. Perelló, J. Palau and J. A. Subirana.
Eur. J. Biochemistry 214:879-887. (1993)
A and Z canonical conformations in d(CnGCGn) crystals characterized by microraman spectroscopies.
H.Sfihi, J. Liquier, L. Urpí, N. Verdaguer, J.A. Subirana, J. Igolen and E. Taillandier
Biopolymers 33:1715-1723. (1993)
Structure of trans-diaquabis[(2-oxopyrrolidin-1-yl)acetamide]copper(II) perchlorate.
X. de la Cruz, A. Martinez-Balbás, J. Tormo and N. Verdaguer.
Acta cryst. C48:167-169. (1992)
Ll.Fajarí, J.Riera, Ll.Juliá ,J.Lloveras, N.Verdaguer and I.Fita.
J.Organometalics Chemistry 423:163-171. (1992)
Molecular structure of a complete turn of A-DNA.
N.Verdaguer, D.Fernández, J.Aymamí, I.Fita, M. Coll T.Huynh-Dinh, J.Igolen and J.A.Subirana
J.Mol. Biol. 221:623-625. (1991)
Molecular Structure of L-Lysyl-L-Tyrosyl-L-Serine Acetate.
N.Verdaguer, I.Fita and J.A.Subira.
Int J Biol Macromol. 12:315-320. (1990)
Crystal and Molecular Structure of L-Lysyl-L-Valine Hydrochloride a new Lysine Conformation.
Ll.Boqué, N.Verdaguer, L.Urpí, I.Fita and J.A.Subirana.
Int.J.Peptide and Protein Res. 33:157-161. (1989)
L.Urpí, J.P.Ridoux, J.Liquier, N.Verdaguer, I.Fita, J.A.Subirana, F.Iglesias, T.Huynh-Dinh, J.Igolen and E.Taillandier.
Nucleic Acids Res. 17:6669-6680. (1989)
Molecular Structure of L-Lysyl-L-Alanyl-L-Alanine. A tripeptide found in Histone H1
N.Verdaguer, L.Urpí, I.Fita, and J.A.Subirana.
Biopolymers. 27:1887-1896. (1988)
Book Chapters
Modificación de la Densidad Electrónica en el Afinamiento y Extensión de las Fases en Macromoléculas.
J. Bravo, N. Verdaguer, I.Fita.
Nuevas Tendencias en Cristalografía. Consejo Superior de Investigaciones Científicas, Madrid, 1995.
Aportaciones de la cristalografía al estudio del ADN.
J.A. Subirana, X. Salas, L.Urpí E. Font, N. Verdaguer.
Nuevas Tendencias en Cristalografía. Consejo Superior de Investigaciones Científicas, Madrid, 1995.
Cristalografía de Macromoléculas
I.Fita, J. Bravo, N. Verdaguer
Simulación Molecular y Ciencias de Materiales. Ed. Bernardo Celda. Universidad Menéndez y Pelayo, Valencia. (1995)
A. Guasch, N.Verdaguer, X.Gomis-Rüth, I.Fita and M.Coll
Dinàmica estructural de Macromolècules. Trb. Soc. Cat. Biol. 48,. Ll.Cornudella Ed. (1999)
Estructura de grandes agregados moleculares: estudios combinados de cristalografia de rayos-X y microscopia electrónica
N. Verdaguer, W.F.Ochoa, D. Blaas, E. Hewat and I. Fita.
Trb. Soc. Cat. Biol. 53 115-124. (2002)
Handbook of Metalloproteins C2 domains
N.Verdaguer, I.Fita, M.Senena-Corbalan, J.C.Gomez-Fernandez
John Wiley & Sons, Ltd. Eds. A. Messerschmidt, R. Huber, T. Poulos and K. Wieghardt. (2003)
Functional and Structural Aspects of the Interaction of Foot-and-Mouth Disease Virus with Antibodies
M.G.Mateu & N. Verdaguer.
In Foot-and-mouth disease current perspectives. Horizon Bioscience. Eds. F. Sobrino and E. Domingo. (2004)
RNA Virus Polymerases. In Viral Genome Replication. II. Elements, factors and enzymes: Structure-function and mechanism.
C. Ferrer-Orta, N. Verdaguer Cameron, Götte and Raney eds.
Springer, New York, (2009)
Mutation, Quasispecies and Lethal Mutagenesis in Picornaviruses.
Domingo E, Perales C, Agudo R, Arias A, Escarmís C, Ferrer-Orta C, Verdaguer N. Ehrenfeld, Domingo and Roos eds.
ASM Press, Washington DC. (2010)
Structural Dynamics of Picornaviral RdRP Complexes. Implications for the Design of Antivirals
Verdaguer N. Ferrer-Orta C, Domingo, E, M.A. Carrondo and P. Spadon (eds)
Springer Science+Business Media B.V. (2012)
Structural Dynamics of the Vault Ribonucleoprotein Particle
Casañas A, Querol J. Fita I, Verdaguer N, M.A. Carrondo and P. Spadon (eds.)
Springer Science+Business Media B.V. (2012)
Virus, el eslabón perdido entre organismos vivos y el mundo inerte. Libro: A través del cristal: como la cristalografía ha cambiado la visión del mundo.
Ferrer-Orta C, Querol-Audí J & Verdaguer N.
Colección Divulgación. CSIC, ISBN: 978-84-00-09800-1. (2014)
Project funding
Ongoing Projects
PID2020-117976GB-I00 Title: “The role of protein-protein and protein-nucleic acid interactions in viral infection”. From September 1, 2021 to August 31, 2024
Amount € 278.300
Role: PI
Ministerio de Ciencia e Innovación
Proyecto PID2020-117976GB-I00 financiado por:

PLEC2021-007654 Title: “Nueva inmunoterapia contra el cáncer: bloqueo de la reprogramación lipídica basada en el mapeo de transcriptomas de metástasis (LipIMMUNE)” . From November, 1, 2021 to October 31, 2024
Amount € 190.000
Role: PI
Ministerio de Ciencia e Innovación
Proyecto PLEC2021-007654 financiado por:

Fundación “La Marató de TV3” Ref. 202136-30. Title: Towards understanding the molecular mechanisms of lethal mutagenesis in SARS-CoV-2. From July 30, 2021 to July 29, 2024
Amount € 150.000
Role: PI
“Fundació La Marató de TV3” Research Project, Edition 2021 –Research Projects in COVID-19
CSIC Salud Gobal PTI+ Plataforma de Antivirales. Title: “Mutagénesis letal sinérgica para SARS-CoV-2 con muestras de virus de pacientes” From Jan 1, 2022 to December 31, 2022
Amount € 78200
Role: co-PI
CSIC
Reference: 201833-10
Title: De l’estructura als mecanismes pels quals el virus Zika provoca neuropaties congènites
Total awarded: €160.000 (30.5.2019 – 30.12.2022)
Role: PI
“Fundació La Marató de TV3” Research Project, Edition 2017 – Biomedical Research Projects in Infectious Diseases
Concluded Projects
Reference: BIO2017-83906-P
Title: Bases estructurales de la infecion viral: Maquinarias replicativas de virus RNA
Total awarded: € 260.000 (1.1.2018 – 31.08.2022) Role: PI
Ministerio de Economía y Competitividad
Proyecto BIO2017-83906-P financiado por:

Reference: MDM-2014-0435
Title: Appointment of the Department of Structural Biology of IBMB as a “María de Maeztu Unit of Excellence”
Total awarded: €2,000,000 (1.7.2015 – 31.12.2019)
Spanish Ministry of Economy and Competitivity.

CSIC-COV-19-014. Title: “Combinaciones de antivirales frente a SARS-CoV-2”
From April 1, 2020 to December 31, 2021
Total awarded: € 160.000
CSIC
2020AEP121 Title: “Bases estructurales de la infección viral: interacción entre proteínas virales y proteínas del hospedador”. From Jan 1, 2020 to August 31, 2021
Total awarded: € 43.262
CSIC
Vacancies/Jobs
Lab corner
No items available
Project gallery
No albums or photos uploaded yet.
