Lab presentation
Our research group is focused on using computational and experimental approaches to understand and design protein functions. We have extensive experience in deep unsupervised learning and protein design, which we have applied to various projects (see below!).
Over the next years, we will expand our focus to include the design of custom-tailored and new-to-nature protein functions. This will involve machine learning and other computational approaches to explore unexplored regions of the protein space and generate novel proteins with desired functions. We will also include experimental characterization efforts, using various techniques to validate and refine our computational models.
We are particularly interested in using our expertise to address significant challenges in the fields of healthcare and sustainability. This includes developing new drugs to treat diseases, designing enzymes for biotechnological applications, and creating proteins with novel functions that can help address environmental challenges.
We believe that protein design has the potential to change the world we live in, and Artificial Intelligence is at the core of this revolution.
Projects
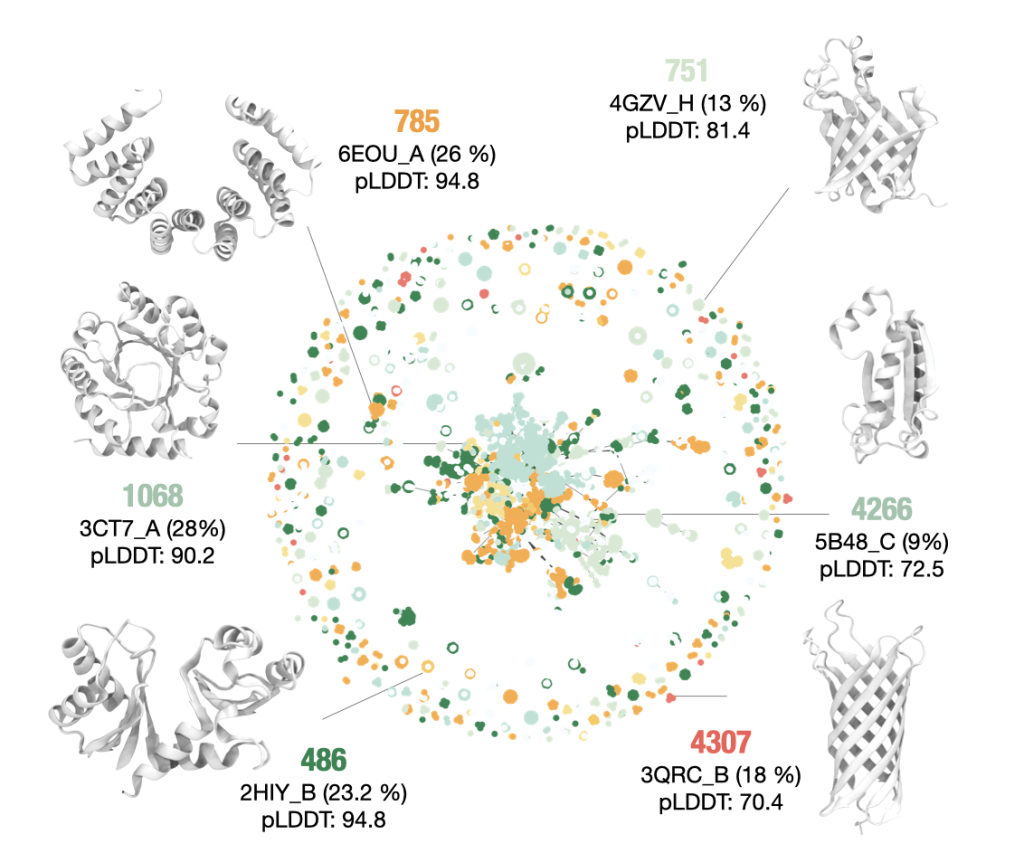
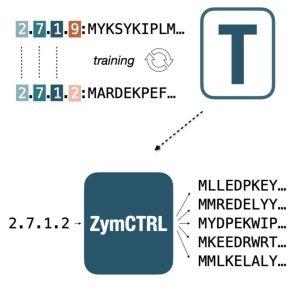
In this project, we trained a GPT2-like architecture to design enzymes with specific functions. Each enzyme sequence was linked to each catalytic identifier (EC class, e.g., ‘2.7.1.2’); hence, the model has learned to map sequence features specific to each enzymatic function. ZymCTRL generates enzyme sequences upon a user-defined catalytic activity prompt.
Munsamy, M., Lindner, S, Lorenz, P., Ferruz, N. ZymCTRL: A conditional model for the controllable generation of artificial enzymes. in the MLSB workshop of the 36th NeurIPS conference (2022). Read preprint
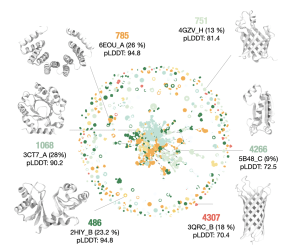
Natural Language Processing methods have shown impressive capabilities generating long, coherent text (think GPT3 or ChatGPT). Inspired by this success, we trained ProtGPT2 on the entire protein space. ProtGPT2 has learned ‘the protein language’ and generates protein sequences in unexplored regions of the protein space. The generated proteins are ordered, globular and feature non-idealized structures, while expressing in wet-lab settings.
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat Commun 13, 4348 (2022). https://doi.org/10.1038/s41467-022-32007-7
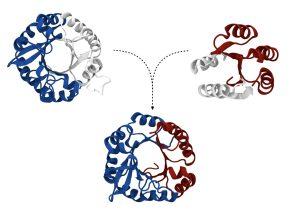
Ferruz, N. et al. Identification and Analysis of Natural Building Blocks for Evolution-Guided Fragment-Based Protein Design. J. Mol. Biol. 432, 3898–3914 (2020).
Ferruz, N., Noske, J. & Höcker, B. Protlego: A Python package for the analysis and design of chimeric proteins. Bioinformatics. 37, 3182–3189 (2021). Read main publication
Lab people

Noelia Ferruz Capapey

Ramiro Illanes Vicioso

Núria Mimbrero

Joan Coines
Selected publications
Deep Learning
Munsamy, G., Lindner, S., Lorenz, P., Ferruz, N. ZymCTRL: a conditional language model for the controllable generation of artificial enzymes
Ferruz, N. et al. From sequence to function through structure: deep learning for protein design. Comput. Struct. Biotechnol. J. (2022) doi:10.1016/j.csbj.2022.11.014.
Ferruz, N. & Höcker, B. Controllable protein design with language models. Nat. Mach. Intell. 4, 521–532 (2022). doi: 10.1038/s42256-022-00499-z
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat. Commun. 13, 4348 (2022). doi: 10.1038/s41467-022-32007-7
Ferruz, N. & Höcker, B. Dreaming ideal protein structures. Nat. Biotechnol. 40, 171–172 (2022). doi: 10.1038/s41587-021-01196-9
Evolutionarily Conserved Fragments
Ferruz, N., Schmidt, S. & Höcker, B. ProteinTools : a toolkit to analyze protein structures. Nucleic Acids Res. 49, W559–W566 (2021). doi: 10.1093/nar/gkab375
Ferruz, N., Michel, F., Lobos, F., Schmidt, S. & Höcker, B. Fuzzle 2.0: Ligand Binding in Natural Protein Building Blocks. Front. Mol. Biosci. 8, (2021). doi: 10.3389/fmolb.2021.715972
Ferruz, N., Noske, J. & Höcker, B. Protlego: A Python package for the analysis and design of chimeric proteins. Bioinforma. Oxf. Engl. 37, 3182–3189 (2021). doi: 10.1093/bioinformatics/btab253
Ferruz, N. et al. Identification and Analysis of Natural Building Blocks for Evolution-Guided Fragment-Based Protein Design. J. Mol. Biol. 432, 3898–3914 (2020). doi: 10.1016/j.jmb.2020.04.013
Large-scale Molecular Dynamics
Kröger, P., Shanmugaratnam, S., Ferruz, N., Schweimer, K. & Höcker, B. A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF. Struct. Lond. Engl. 1993 29, 433-443.e4 (2021). doi: 10.1016/j.str.2020.12.005
Ferruz, N. et al. Dopamine D3 receptor antagonist reveals a cryptic pocket in aminergic GPCRs. Sci. Rep. 8, 897 (2018). doi: 10.1038/s41598-018-19345-7
Ferruz, N., Tresadern, G., Pineda-Lucena, A. & De Fabritiis, G. Multibody cofactor and substrate molecular recognition in the myo-inositol monophosphatase enzyme. Sci. Rep. 6, 30275 (2016). doi: 10.1038/srep30275
Ferruz, N., Harvey, M. J., Mestres, J. & De Fabritiis, G. Insights from Fragment Hit Binding Assays by Molecular Simulations. J. Chem. Inf. Model. 55, 2200–2205 (2015). doi: 10.1021/acs.jcim.5b00453
Lauro, G. et al. Reranking Docking Poses Using Molecular Simulations and Approximate Free Energy Methods. J. Chem. Inf. Model. 54, 2185–2189 (2014). doi: 10.1021/ci500309a
All publications
Deep Learning
Munsamy, G., Lindner, S., Lorenz, P., Ferruz, N. ZymCTRL: a conditional language model for the controllable generation of artificial enzymes
Ferruz, N. et al. From sequence to function through structure: deep learning for protein design. Comput. Struct. Biotechnol. J. (2022) doi:10.1016/j.csbj.2022.11.014.
Ferruz, N. & Höcker, B. Controllable protein design with language models. Nat. Mach. Intell. 4, 521–532 (2022). doi: 10.1038/s42256-022-00499-z
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat. Commun. 13, 4348 (2022). doi: 10.1038/s41467-022-32007-7
Ferruz, N. & Höcker, B. Dreaming ideal protein structures. Nat. Biotechnol. 40, 171–172 (2022). doi: 10.1038/s41587-021-01196-9
Evolutionarily Conserved Fragments
Ferruz, N., Schmidt, S. & Höcker, B. ProteinTools : a toolkit to analyze protein structures. Nucleic Acids Res. 49, W559–W566 (2021). doi: 10.1093/nar/gkab375
Ferruz, N., Michel, F., Lobos, F., Schmidt, S. & Höcker, B. Fuzzle 2.0: Ligand Binding in Natural Protein Building Blocks. Front. Mol. Biosci. 8, (2021). doi: 10.3389/fmolb.2021.715972
Ferruz, N., Noske, J. & Höcker, B. Protlego: A Python package for the analysis and design of chimeric proteins. Bioinforma. Oxf. Engl. 37, 3182–3189 (2021). doi: 10.1093/bioinformatics/btab253
Ferruz, N. et al. Identification and Analysis of Natural Building Blocks for Evolution-Guided Fragment-Based Protein Design. J. Mol. Biol. 432, 3898–3914 (2020). doi: 10.1016/j.jmb.2020.04.013
Large-scale Molecular Dynamics
Kröger, P., Shanmugaratnam, S., Ferruz, N., Schweimer, K. & Höcker, B. A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF. Struct. Lond. Engl. 1993 29, 433-443.e4 (2021). doi: 10.1016/j.str.2020.12.005
Ferruz, N. et al. Dopamine D3 receptor antagonist reveals a cryptic pocket in aminergic GPCRs. Sci. Rep. 8, 897 (2018). doi: 10.1038/s41598-018-19345-7
Ferruz, N., Tresadern, G., Pineda-Lucena, A. & De Fabritiis, G. Multibody cofactor and substrate molecular recognition in the myo-inositol monophosphatase enzyme. Sci. Rep. 6, 30275 (2016). doi: 10.1038/srep30275
Ferruz, N., Harvey, M. J., Mestres, J. & De Fabritiis, G. Insights from Fragment Hit Binding Assays by Molecular Simulations. J. Chem. Inf. Model. 55, 2200–2205 (2015). doi: 10.1021/acs.jcim.5b00453
Lauro, G. et al. Reranking Docking Poses Using Molecular Simulations and Approximate Free Energy Methods. J. Chem. Inf. Model. 54, 2185–2189 (2014). doi: 10.1021/ci500309a
Project funding
- Project title: MINECO (PID2022-139006NA-I00) ‘Design of custom-tailored proteins with unsupervised conditional language models (ProtCTRL)’
- Funding source: Spanish Ministry of Science
- Amount: 143.750 €
- Period: 09/2023-08/2026
Proyecto PID2022-139006NA-I00 financiado por:

- Project title: MINECO (CPP2022-009990) ‘ARTIFICIAL INTELLIGENCE-BASED DESIGN OF GENE WRITERS FOR ADVANCED THERAPIES’
- Funding source: Spanish Ministry of Science
- Amount: 332.463,8 €
- Period: 11/2023-10/2026
- Project title: BBVA Leonardo Creators – ‘Language translation models for on-demand protein generation’
- Funding source: Banco Bilbao Vizcaya Argentaria (BBVA)
- Amount: 39.850 €
- Period: 2024
- Project title: ‘Explainable AI for molecules – AIChemist’ Marie Skłodowska-Curie Actions. Industrial Doctoral Networks. (GA: 101120466)
- Funding source: European Research Council
- Amount: 125.985 (1 PhD student)
- Period: 2023-2027
Project AiChemist (101120466) funded by (HORIZONTE EUROPA, HORIZON-MSCA-2022-DN-01).

- Project title: ‘Screening and Selection of Multivalent Immune Checkpoint Inhibitory Peptides (ImCheScreen- V9172’ SCREENTECH)
- Funding source: Spanish Ministry of Science/Catalan Government
- Amount: 56.500 €
- Period: 2024-2025
- Project title: Starting Package ‘Ramon y Cajal Fellowship. (RYC2021-034367-I)
- Funding source: Spanish Ministry of Science
- Amount: 52.500 €
- Period: 2024- 2025
- Reference: RYC2021-034367-I
- Amount: 305.777,56 €
- Dates: from 01/01/2023 to 31/12/2027
- Principal Investigator: Noelia Ferruz
Ayuda RYC2021-034367-I financiada por:

Vacancies/Jobs
We are always seeking motivated researchers at all career stages to join the lab. Please, have a look at our webpage for more specific openings or send an email to Noelia Ferruz (nfccri@ibmb.csic.es) for more details.
Lab corner
Project gallery
Contact

