Lab presentation
Protein Design and Modeling
Proteins are extraordinarily versatile biomolecules that carry out a broad range of functions in life – including molecular recognition, catalysis, transport and scaffolding – and uniquely inspire us for engineering molecular solutions to current challenges in therapeutics, synthetic biology, green chemistry, nanomaterials and, more generally, biological research. Our current understanding of how protein sequences dictate three-dimensional structures now enables us to tackle the reverse challenge: designing from first principles protein sequences folding into desired structures, also known as “de novo protein design”. The need for designing and tailoring proteins from scratch is clear: repurposing natural proteins leads to suboptimal or no results in many applications. Despite their proficiency, natural proteins generally suffer from a series of limitations when repurposed for new functions: instability, not having the desired geometry or size, too low recombinant expression levels for large-scale production, exhibiting undesired interactions with other proteins or folding through too complex processes, among others. The de novo protein design revolution has arrived to transform the way we engineer proteins; opening the door to a universe of hyperstable proteins designable with high structural accuracy and enough complexity for tailoring functions of medical and biotechnological relevance.
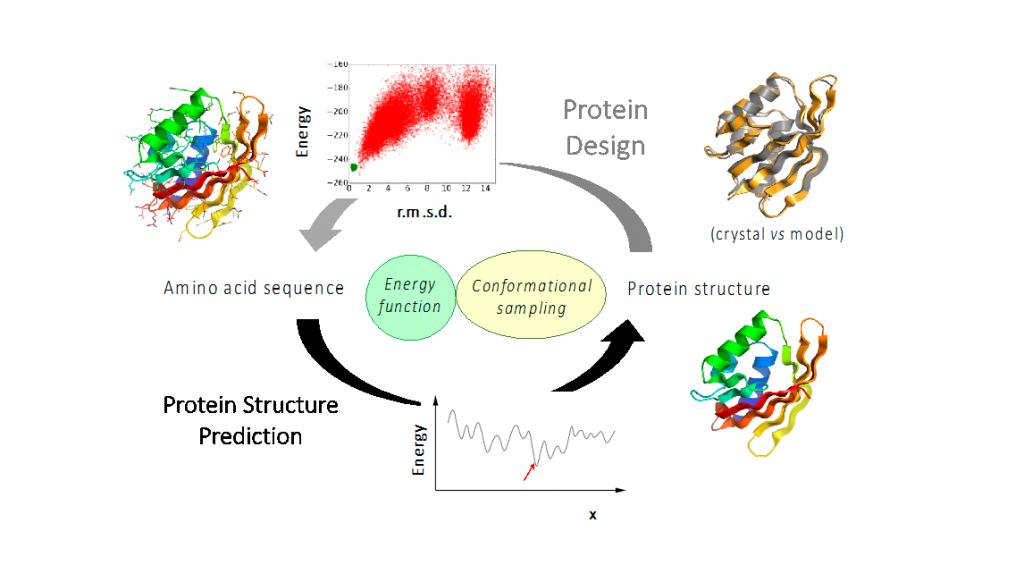
Figure legend. Protein design and structure prediction are two opposite and synergizing problems favoring the development of energy functions and conformational sampling strategies satisfying the principle that the structure of well-folded proteins corresponds to the global energy minima of their amino acid sequence.
Our goal in the Protein Design and Modeling Lab is to combine computational methods and experimental characterization to design brand new proteins for biotechnological and medical applications. We believe that computation and experiment together make the whole design process more efficient, exciting and rewarding. We start by computationally designing proteins using the Rosetta macromolecular modelling software – we belong to the Rosetta Commons – and validate their sequence-structure compatibility with structure prediction simulations. Then, to experimentally verify that the designs behave as predicted, we recombinantly express genes encoding for the designed sequences and characterize the proteins by combining biochemical and biophysical techniques (circular dichroism, MALS…) with X-ray crystallography. Our multidisciplinary research aims at developing design principles and expanding computational protein design methodologies for novel protein folds, binders to small-molecules or proteins, as well as conformational protein switches responding to signals. We are also interested in how computational design pipelines can be improved to increase their predictive power and minimize experimental screening. Towards this goal, we are exploring molecular dynamics simulations and enhanced sampling techniques as stringent validation tests of designer proteins in silico. Our vision is to make de novo protein design a robust approach that is widely accessible to many laboratories and industrial companies interested in different areas of biology and biotechnology.
Major advances in de novo protein design made in the last decade have resulted in a burst of de novo proteins in manifold geometries with high stability and experimental structures matching their computational models. We have contributed to a number of studies adding to de novo proteins the complexity necessary for ligand-binding functions.
We have described the first de novo designed proteins made only with beta-sheets, which are attractive frameworks for enzymes, small-molecule binders and antibodies. Despite more than 20 years of protein design research, the design of all-beta proteins had remained elusive due to their higher structural complexity and aggregation propensity. We have identified design principles for designing well-packed all-beta proteins using long-range loop connections between beta-strands, and computationally designed double-stranded beta-helices with the jelly-roll fold at atomic-level accuracy. This study has opened the door to custom-design a broad range of beta-sheet proteins suited for binding sites, including de novo beta-sandwiches with the immunoglobulin fold as novel antibody frameworks.
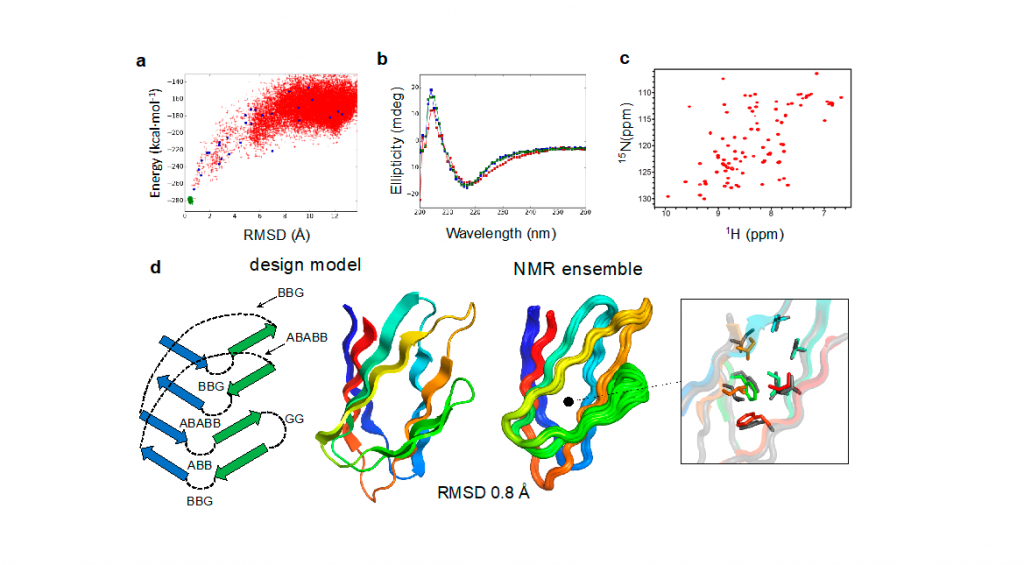
Figure Legend. De novo design of a double stranded beta-helix. (a) Computationally designed sequences with funnel-shaped energy landscapes were then expressed in E coli and (b) their secondary structure and thermostability was confirmed with circular dichroism. (c) The formation of well-folded structures was verified by nuclear magnetic resonance HSQC. (d) The structure of the best design model was solved by nuclear magnetic resonance in excellent agreement with the computational model.
Based on structural analysis of natural proteins we have identified the mechanisms by which natural proteins bend beta-sheets when building binding pockets, and then developed a computational approach for de novo designing NTF2-like protein folds with cavities controlled by curved beta-sheets, which are ideal protein scaffolds for tailoring small-molecule binding sites in biosensing and biocatalysis applications.
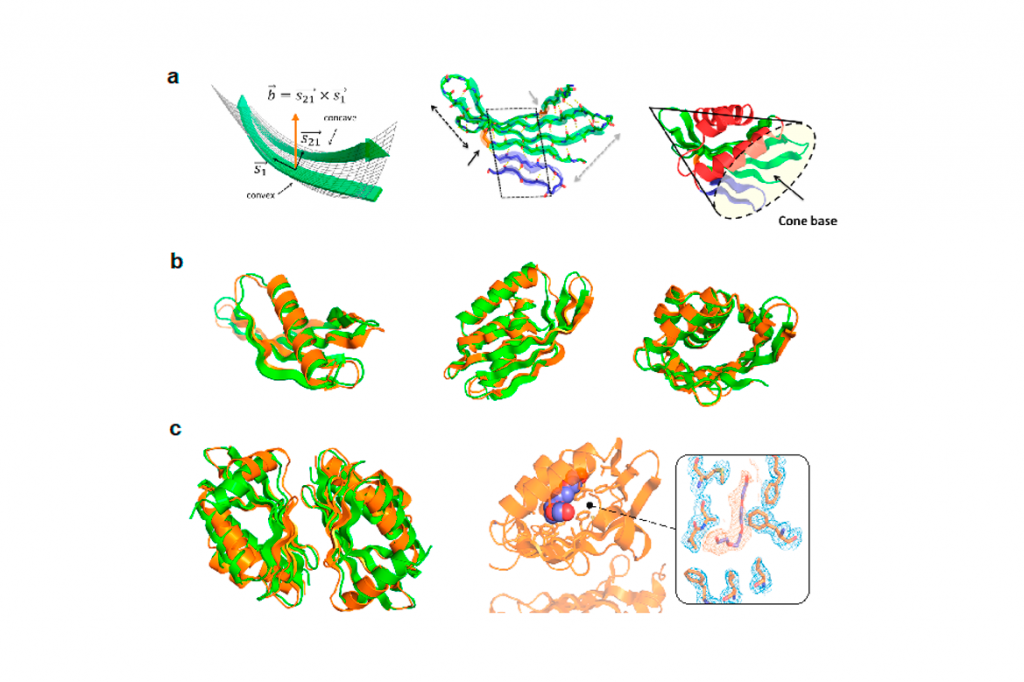
Figure Legend. De novo design of proteins with cavities controlled by curved beta-sheets. (a) Analysis of beta-sheet curvature and identification of topological parameters controlling beta-sheet geometry and that were used to build cone-like structures for hosting cavities. (b) Examples of designed proteins with X-ray or NMR structures matching their computational models. (c) Homodimers designed for enhancing stability and growing buried cavities able to bind small-molecules.
Projects
- De novo design of beta-rich protein folds suited for small-molecule or protein binding.
- De novo design of novel protein folds with increasingly complex and irregular structural features necessary for function.
- De novo design of protein conformational switches.
- Computational design of proteins blocking pathogenic proteins.
- Molecular dynamics screening of computationally designed proteins.
- De novo design of non-local beta-sheet protein scaffolds (PID2020-120098GA-100). Proyectos I+D+I 2020. Ministerio de Ciencia e Innovacion. IBMB-CSIC. 2021-2024
Lab people

Enrique Marcos Benteo
Dr. Enrique Marcos graduated in Chemistry from the Instituto Químico de Sarriá (Universidad Ramon Llull) of Barcelona in 2005. He then moved to the Instituto de Química Avanzada de Cataluña (IQAC/CSIC) to carry out his predoctoral studies on molecular modelling of enzymes under the supervision of Dr. Ramon Crehuet. During that time he visited the labs of Dr. Ivet Bahar at University of Pittsburgh and Dr. Martin Field at the IBS in Grenoble. In 2012 he obtained his PhD in Computational Chemistry and then, from 2012 to 2015 as a Marie Curie Outgoing postdoctoral fellow, joined the group of Dr. David Baker at the University of Washington, Seattle, United States. During that time he worked on the de novo design of protein folds suited for ligand-binding functions. With a Marie Curie Reintegration fellowship he then came back and joined IRB Barcelona under the supervision of Prof. Modesto Orozco. In 2019 he was awarded a Junior Leader Retaining grant from “La Caixa” to start his independent career, and then in 2020 has joined IBMB/CSIC as a “Ramon y Cajal” researcher to build a multidisciplinary research line on protein design.

Marta Nadal Rovira

Daniel Peñas Utrilla

Lourdes Carcelén

Alexandre Casadesus
Past Students
Jorge Luis Roel Touris
Nicolás Byrne Álvarez
Selected publications
Roel-Touris, J., Nadal, M. & Marcos, E. Single-chain dimers from de novo immunoglobulins as robust scaffolds for multiple binding loops. Nat Commun 14, 5939 (2023). https://doi.org/10.1038/s41467-023-41717-5
Peñas-Utrilla D and Marcos E (2022) Identifying well-folded de novo proteins in the new era of accurate structure prediction. Front. Mol. Biosci. 9:991380. doi: 10.3389/fmolb.2022.991380
Chidyausiku, T.M., Mendes, S.R., Klima, J.C., Gomis-Rüth, F. Xavier, Marcos, Enrique et al. De novo design of immunoglobulin-like domains. Nat Commun 13, 5661 (2022). https://doi.org/10.1038/s41467-022-33004-6
Silva DA, Yu S, Ulge U, Spangler J, Jude K, Labao-Almeida C, Ali L, Quijano-Rubio A, Ruterbusch M, Leung I, Biary T, Crowley S, Marcos E, Walkey C, Weitzner B, Pardo-Ávila F, Castellanos J, Carter L, Stewart L, Riddell S, Pepper M, Bernades G, Dougan M, Garcia C, Baker D*. “De novo design of potent and selective mimics of IL-2 and IL-15”, Nature, 565, 186-191 (2019). doi: 10.1038/s41586-018-0830-7
Marcos E*, Chidyausiku TM, McShan A, Evangelidis T, Nerli S, Carter L, Nivon L, Davis A, Oberdorfer G, Tripsianes K, Sgourakis N, Baker D*. “De novo design of a non-local β-sheet protein with high stability and accuracy”, Nature Structural & Molecular Biology, 25, 1028-1034 (2018). doi: 10.1038/s41594-018-0141-6
Marcos E*, Silva DA*. “Essentials of de novo protein design: Methods and applications”, WIRES Computational Molecular Science, e1374 (2018). doi: 10.1002/wcms.1374
Dou J, Vorobieva A, Sheffler W, Doyle L, Park H, Bick M, Mao B, Foight G, Lee MY, Gagnon L, Carter L, Sankaran B, Ovchinnikov S, Marcos E, Huang PS, Vaughan J, Stoddard B, Baker D. “De novo design of a fluorescent-activating β-barrel”, Nature, 561, 485-491 (2018). doi: 10.1038/s41586-018-0509-0
Marcos E, Basanta B, Chidyausiku TM, Tang Y, Oberdorfer G, Liu G, Swapna GVT, Guan R, Silva DA, Dou J, Pereira JH, Xiao R, Sankaran B, Zwart PH, Montelione GT and Baker D*. “Principles for designing proteins with cavities formed by curved β-sheets”, Science, 355, 201-206 (2017). doi: 10.1126/science.aah7389
Marcos E, Sanchez-Martinez M and Crehuet R. “Interplay between Enzyme Function and Protein Dynamics: a Multiscale Approach to the Study of the NAG Kinase Family and Two Class II Aldolases”, in Computational Approaches to Protein Dynamics: From Quantum to Coarse-Grained Methods, Monika Fuxreiter ed., CRC Press (2015). doi: 10.1201/b17979-11
All publications
Roel-Touris, J., Nadal, M. & Marcos, E. Single-chain dimers from de novo immunoglobulins as robust scaffolds for multiple binding loops. Nat Commun 14, 5939 (2023). https://doi.org/10.1038/s41467-023-41717-5
Peñas-Utrilla D and Marcos E (2022) Identifying well-folded de novo proteins in the new era of accurate structure prediction. Front. Mol. Biosci. 9:991380. doi: 10.3389/fmolb.2022.991380
Chidyausiku, T.M., Mendes, S.R., Klima, J.C., Gomis-Rüth, F. Xavier, Marcos, Enrique et al. De novo design of immunoglobulin-like domains. Nat Commun 13, 5661 (2022). https://doi.org/10.1038/s41467-022-33004-6
Koehler J*, Weitzner B, Lewis S, …, Marcos E, …, Bonneau R*. “Macromolecular Modeling and Design in Rosetta: New Methods and Frameworks”. Nature Methods, 17, 665-680 (2020). doi: 10.1038/s41592-020-0848-2
Koehler J*, Weitzner B, Renfrew D, Lewis S, Moretti R, Watkins A, Mulligan V, Lyskov S, Adolf-Bryfogle J, Labonte J, Krys J, …, Marcos E, …, Bystroff C, Schief W, Gront D, Schueler-Furman O, Baker D, Bradley P, Dunbrack R, Strauss S, Leaver-Fay A, Kortemme T, Meiler J, Kuhlman B, Gray J, Bonneau R*. “Better together: Elements of successful scientific software development in a distributed collaborative community”. PLoS Computational Biology 16(5): e1007507 (2020). doi: 10.1371/journal.pcbi.1007507
Silva DA, Yu S, Ulge U, Spangler J, Jude K, Labao-Almeida C, Ali L, Quijano-Rubio A, Ruterbusch M, Leung I, Biary T, Crowley S, Marcos E, Walkey C, Weitzner B, Pardo-Ávila F, Castellanos J, Carter L, Stewart L, Riddell S, Pepper M, Bernades G, Dougan M, Garcia C, Baker D*. “De novo design of potent and selective mimics of IL-2 and IL-15”, Nature, 565, 186-191 (2019). doi: 10.1038/s41586-018-0830-7
Marcos E*, Chidyausiku TM, McShan A, Evangelidis T, Nerli S, Carter L, Nivon L, Davis A, Oberdorfer G, Tripsianes K, Sgourakis N, Baker D*. “De novo design of a non-local β-sheet protein with high stability and accuracy”, Nature Structural & Molecular Biology, 25, 1028-1034 (2018). doi: 10.1038/s41594-018-0141-6
Marcos E*, Silva DA*. “Essentials of de novo protein design: Methods and applications”, WIRES Computational Molecular Science, e1374 (2018). doi: 10.1002/wcms.1374
Dou J, Vorobieva A, Sheffler W, Doyle L, Park H, Bick M, Mao B, Foight G, Lee MY, Gagnon L, Carter L, Sankaran B, Ovchinnikov S, Marcos E, Huang PS, Vaughan J, Stoddard B, Baker D. “De novo design of a fluorescent-activating β-barrel”, Nature, 561, 485-491 (2018). doi: 10.1038/s41586-018-0509-0
Marcos E, Basanta B, Chidyausiku TM, Tang Y, Oberdorfer G, Liu G, Swapna GVT, Guan R, Silva DA, Dou J, Pereira JH, Xiao R, Sankaran B, Zwart PH, Montelione GT and Baker D*. “Principles for designing proteins with cavities formed by curved β-sheets”, Science, 355, 201-206 (2017). doi: 10.1126/science.aah7389
Torrent-Sucarrat M*, Navarro S, Marcos E, Anglada JM and Luis JM. “Design of Hückel–Möbius Topological Switches with High Nonlinear Optical Properties”, The Journal of Physical Chemistry C, 121 (35), 19348-19357 (2017). doi: 10.1021/acs.jpcc.7b05900
Marcos E*, Anglada JM and Torrent-Sucarrat M*. “Effect of the Meso-Substituent in the Hückel-to-Möbius Topological Switches”, The Journal of Organic Chemistry, 79, 5036-5046 (2014). doi: 10.1021/jo500569p
Sanchez-Martinez M, Marcos E, Tauler R, Field MJ and Crehuet R*. “Conformational Compression and Catalytic Heterogeneity in the N-acetylglutamate Kinase”, The Journal of Physical Chemistry B, 117, 14261-14272 (2013). doi: 10.1021/jp407016v
Marcos E*, Anglada JM and Torrent-Sucarrat M*. “Theoretical Study of the Switching between Hückel and Möbius Topologies for Expanded Porphyrins”, The Journal of Physical Chemistry C, 116 (45), 24358-24366 (2012). doi: 10.1021/jp3083612
Marcos E, Jimenez A and Crehuet R*. “Dynamic Fingerprints of Protein Thermostability Revealed by Long Molecular Dynamics”, Journal of Chemical Theory and Computation, 8, 1129-1142 (2012). doi: 10.1021/ct200877z
Marcos E, Mestres P and Crehuet R*. “Crowding induces differences in the diffusion of thermophilic and mesophilic proteins: a new look at neutron scattering results”, Biophysical Journal, 101, 2782-2789 (2011). doi: 10.1016/j.bpj.2011.09.033
Marcos E, Crehuet R* and Bahar I*. “Changes in dynamics upon oligomerization regulate substrate binding and allostery in Amino Acid Kinase family members”, PLoS Computational Biology, 7: e1002201 (2011). doi: 10.1371/journal.pcbi.1002201
Marcos E, Field MJ and Crehuet R*. “Pentacoordinated phosphorus revisited by high-level QM/MM calculations”, Proteins: Structure Function Bioinformatics, 78, 2405-2411 (2010). doi: 10.1002/prot.22758
Marcos E, Crehuet R* and Bahar I*. “On the conservation of the slow conformational dynamics within the amino acid kinase family: NAGK the paradigm”, PLoS Computational Biology, 6: e100738 (2010). doi: 10.1371/journal.pcbi.1000738
Marcos E, Anglada JM and Crehuet R. “Description of pentacoordinated phosphorus under an external electric field: which basis sets and semi-empirical methods are needed?”, Physical Chemistry Chemical Physics, 10, 2442-2450 (2008). doi: 10.1039/B719792F
Marcos E, Crehuet R* and Anglada J.M*. “Inductive and external electric field effects on pentacoordinated phosphorus compounds”, Journal of Chemical Theory and Computation, 4, 49-63 (2008). doi: 10.1021/ct700220z
Book chapters
• Marcos E, Sanchez-Martinez M and Crehuet R. “Interplay between Enzyme Function and Protein Dynamics: a Multiscale Approach to the Study of the NAG Kinase Family and Two Class II Aldolases”, in Computational Approaches to Protein Dynamics: From Quantum to Coarse-Grained Methods, Monika Fuxreiter ed., CRC Press (2015). doi: 10.1201/b17979-11
Patents
• Chidyausiku TM, Marcos E, Nivon L, Oberdorfer G, Baker D, Carter L. “De Novo Designed JellyRolls aka Cupins Non-Local Beta Sheet Proteins”. Provisional patent application by University of Washington. Registered in United States (27th May 2019) under application number 48546. Patent protection only in United States.
Project funding
Ongoing Projects
Reference: RYC2018-025295-ITitle: Ramon y Cajal research grant, Edition 2018
Total awarded: €40000 (1.9.2020 – 31.8.2025)
Role: PI
Ministerio de Ciencia, Innovación y Universidades
Ayuda RYC2018-025295-I financiada por:

Reference: EUR2020-112164
Title: Diseño de novo de proteinas intercambiantes entre estados conformacionales definidos
Total awarded: €60000 (1.12.2020 – 30.11.2022)
Role: PI
Acciones de dinamización “Europa Excelencia”
Ministerio de Ciencia, Innovación y Universidades
Ayuda EUR2020-112164 financiada por:

Concluded Projects
Reference: LCF/BQ/PR19/11700006Title: Junior Leader Retaining research grant, Edition 2019
Total awarded: €51333 (1.5.2019 – 31.8.2020)
Role: PI
Fundación “La Caixa”
Vacancies/Jobs
We’re building a multidisciplinary and enthusiastic group of protein scientists. We always look for talented undergraduate and graduate students for internships, short-term visits and long-term projects.
Lab corner
Project gallery
No albums or photos uploaded yet.
