Lab presentation
A unique feature of mitochondria is that they possess their own genome, the mitochondrial DNA (mtDNA), which codes for subunits of the electron transport chain that eventually leads to the synthesis of the ATP molecule. Dysregulation of this genome causes complex diseases and syndromes that are difficult to diagnose and treat. These are mostly classified as rare diseases. In the Structural MitoLab, we are interested in the structural basis underlying mtDNA regulation and packaging.
Another important line of research in our group are infectious diseases, which we approach by unveiling the mechanistic basis of the activity of proteins essential for organismal viability.
Our research is multidisciplinary and combines macromolecular X-ray crystallography with biochemical and biophysical analyses (chromatography, electrophoresis, SAXS, MALLS, etc.), which can be completed with additional techniques in collaboration with other groups.
Projects
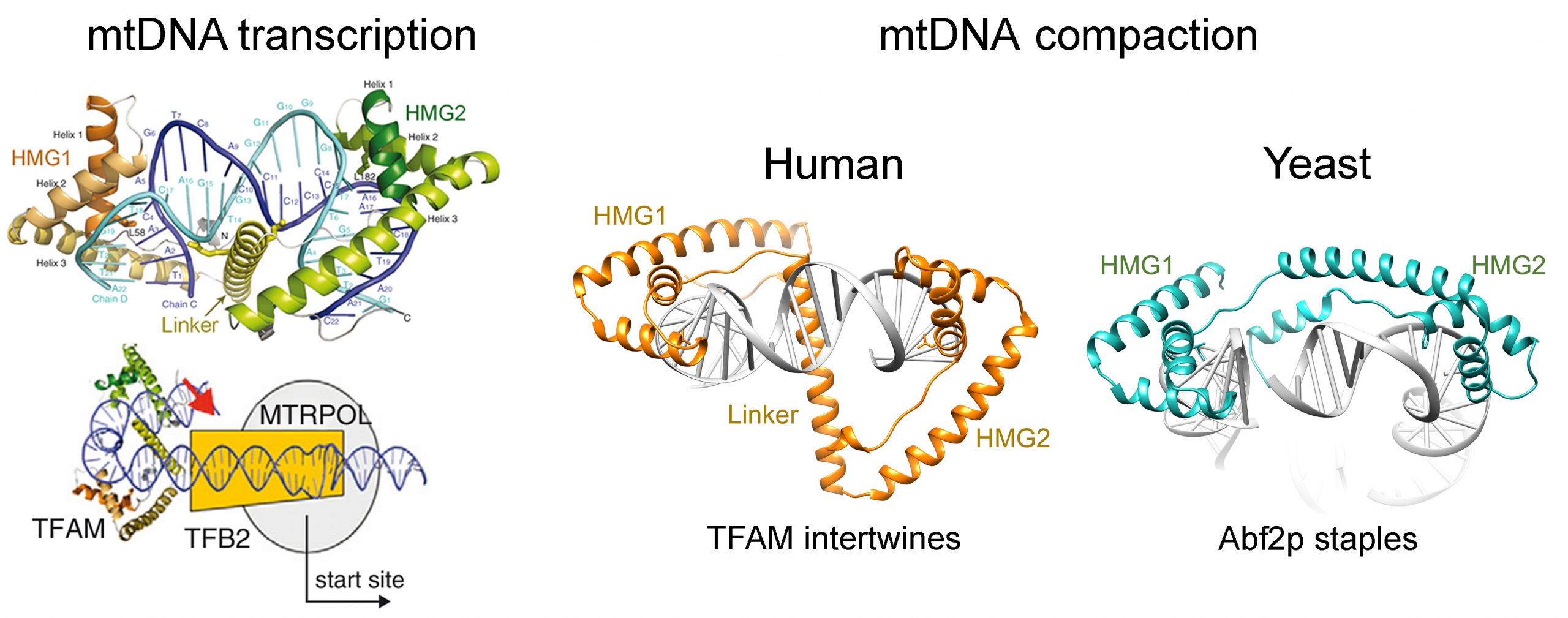
Our group is interested in understanding the structural basis of mitochondrial genome regulation. In humans, the mtDNA is a circular molecule of 16.5kbp that codes for 13 essential subunits (out of approximately 80) of complexes I, III, IV and V of the oxidative phosphorylation pathway (OXPHOS), as well as for two rRNAs and 22tRNAs. The remaining OXPHOS subunits are imported from the cytosol. During mitochondrial fusion and fission, mtDNA is replicated and transmitted to daughter organelles. Therefore, correct transcription, replication and maintenance of the mtDNA are essential for cell viability.
Structural basis of mitochondrial DNA transcription and compaction. The mtDNA transcription initiation complex is formed by mitochondrial transcription factor A (TFAM) binding to one of the two mtDNA transcription promoters, HSP1 or LSP. At these promoters, TFAM recruits the RNA polymerase (mtRNAP). Factor B2, who binds mtRNAP, creates the replication bubble. We solved the crystal structure of TFAM bound to LSP. The protein encompasses two HMG-box domains, which are typically found in the nucleus. In TFAM, these domains insert residues into the DNA, disrupt the stacking between bases and induce a strong bend of 180º (Rubio-Cosials et al., 2011). In the complex, the protein and the DNA are intertwined, a binding mechanism that is possible by virtue of unliganded TFAM’s high flexibility and cooperativity between domains, as we showed by SAXS and FRET (Rubio-Cosials et al., 2011 and 2017). Two apparently homologous TFAM binding sites, DNA Site-X and Site-Y, similar to and downstream from LSP, unexpectedly show distinct DNA-structure features and TFAM binds to them in opposite directions, imposing again a U-turn (Cuppari el al., 2019). In these studies we showed that TFAM/DNA complexes multimerize at higher concentrations. Our biochemical analyses showed that TFAM binds to DNA structures, such as G-quadruplexes (Lyonnais et al., 2017).
All the data above posed the question whether a U-turn is the mtDNA compaction unit.
The U-turn was also found in the complex of S. cerevisiae mtDNA compacting protein Abf2p (Chakraborty et al., 2017). This protein also bears two HMG-boxes but the mechanism of binding is from one side of the DNA, as a staple. Therefore, the fact that evolutionary-distant eukaryotes show a similar U-turn suggested that this is the universal compaction unit, induced by different mechanisms.
Proteins involved in mtDNA replication. Other proteins studied in the laboratory and whose structure we solved include single-strand binding protein mtSSB (or SSBP1, Piro-Mégy et al., 2020), the replicative helicase Twinkle (Fernández-Millán et al, 2015), and the transcription attenuator mTERF. Twinkle and mtSSB were, in addition of being observed in mitochondria, found in the RNE granules that lay close to the mtDNA (Hensen et al., 2019)
Proteins from pathogens, the way to a cure:
The laboratory is interested in the determination of the structure of proteins important for pathogen viability and virulence. This includes the peptidylarginine deiminase PPAD from the widespread odontopathogen Porphyromonas gingivalis. PPAD deiminates the guanidinium group of arginine, leading to excessive citrullination thereby causing inflammation. PPAD was solved with a general PAD inhibitory molecule at the active site. Furthermore, we also participated in the structure of a peptidase and sortase PorU, which processes virulent factors that are translocated by the T9 secretion system through the outer membrane. The results showed that PorU latency is in the form of a dimer while the active form as a monomer.
Lab people

Maria Solà Vilarrubias
The laboratory is headed by Dr. Maria Solà. After a tenure-tracked period of four years funded by a Ramón y Cajal contract, Maria Solà was awarded a permanent position as tenured Assistant Professor at IBMB-CSIC in 2008 and promoted to Associate Professor in 2019.
The research of the group, dedicated to proteins of biomedical interest, including proteins essential for mitochondrial or pathogen viability, involves post-doctoral fellows, PhD candidates and master students. National and international collaborations provide interdisciplinarity and give an expanded vision in the analysis of the proteins of interest.
Maria Solà also participates in teaching at courses and in scientific dissemination events.
ORCID: ORCID 0000-0002-0838-9706
Projects:
R&D project, Societal Challenges State Plan, Life Sciences Subplan, Ministry of Science, Innovation and Universities. “Structural analysis of proteins essential for mitochondria and pathogen viability”. Ref.: RTI2018-101015-B-100. Role: PI. 1.1.2019-31.08.2022.

Elena Ruiz

Nord Oliver Turro

Mauricio Ernesto Orozco Ugarriza
Past students
Claus Schmitz
Aleix Tarres Sole
Arkadiusz Szura
María Rebollo de Sandoval
Selected publications
Spínola-Amilibia, M., Illanes-Vicioso, R., Ruiz-López, E., Colomer-Vidal, P., Ventura Rodriguez, F., Peces Pérez, R., Arias, C.F., Torroba, T., Solà, M.*, Arias-Palomo, E.*, Bertocchini, F.* (2023). Plastic degradation by insect hexamerins: Near-atomic resolution structures of the polyethylene-degrading proteins from the wax worm saliva. Science Advances 9, eadi6813, DOI: 10.1126/sciadv.adi6813
Tarrés-Solé, A., Battistini, F., Gerhold, J.M., Piétrement, O., Martínez-García, B., Ruiz-López, E., Lyonnais, S., Bernadó, P., Roca, J., Orozco, M., Le Cam, E., Sedman, J. & Solà, M.* (2023). Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism. Nucleic Acids Research, Volume 51 (11), Pages 5864–5882, doi.org/10.1093/nar/gkad397
Schmitz, C., Madej, M*., Nowakowska, Z., Cuppari A, et al., Potempa J *, Solà M* (2022). Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors. Nucleic Acids Research, Volume 50 (21), pp. 12558- 12577, doi.org/10.1093/nar/gkac1103
Mizgalska, D., Goulas, T, Rodríguez-Banqueri, A., Veillarda, F., Madej, M., Małeckaa, E., Szczesniaka, K., Ksiazeka. M., Widziołeka, M., Guevara, T., Eckhardb, U., Solà, M.*, Potempa,J.*, Gomis-Rüth, F.-X.* (2021). Intermolecular latency regulates the essential C-terminal signal peptidase and sortase of the Porphyromonas gingivalis type-IX secretion system. PNAS 2021 Vol. 118 No. 40 e2103573118. DOI: 10.1073/pnas.2103573118
Piro-Mégy, C., Sarzi, E., Tarrés-Solé, A., Péquignot, M., Hensen, F., Quilès, M., Manes, G., Sénéchal, A., Bocquet, B., Cazevieille, C., Roubertie, A., Müller, A., Charif, M., Goudenège, D., Lenaers, G., Wilhelm, H., Kellner, U., Weisschuh, N., Wissinger, B., Zanlonghi, X., Spelbrink* J.N., Sola, M.*, Delettre, C.* (2020). Dominant mutations in SSBP1 impair mtDNA replication and cause optic atrophy and foveopathy. Journal of Clinical Investigation, 130(1):143-156. DOI: 10.1172/JCI128513
Cuppari, A., Fernández-Millán, P., Battistini, F., Tarrés-Solé, A., Lyonnais, S., Iruela, G., Ruiz-López, E., Enciso, Y., Rubio-Cosials, A., Prohens, R., Pons, M., Alfonso, C., Tóth, K., Rivas, G., Orozco, M., Solà, M. * (2019) DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region. Nucleic Acids Research, 47: 6519–6537. DOI: 10.1093/nar/gkz406
- Selected as “Article of the Month” in the July 2019 issue of the Journal of the Spanish Society of Biophysics (SEB), Biofísica.
Picchioni, D., Antolin-Fontes, A., Camacho, N., Schmitz, C., Pons-Pons, A., Rodriguez-Escribà, M., Machallekidou, Güller, M.N., Siatra, P., A., Carretero-Junquera, M., Serrano, A., Hovde, S.-L., Knobel, P.A., Novoa E., Solà-Vilarrubias, M., Kaguni, L.S., Stracker, T.H., Ribas de Pouplana, L. (2019) Mitochondrial Protein Synthesis and mtDNA Levels Coordinated through an Aminoacyl-tRNA Synthetase Subunit. Cell Reports, 271:40-47. DOI: 10.1016/j.celrep.2019.03.022
F. Hensen, A. Potter, S. Esvel, A. Tarrés-Solé, A. Chakraborty., M. Solà, J.N. Spelbrink* (2019) Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Research 47(7):3680-3698. DOI: 10.1093/nar/gkz047. IF= 11.147 (2018/2019).
Bereta, G., Goulas, T., Madej, M., Bielecka, E., Solà, M.*, Potempa, J.*, Gomis-Rüth, F.X.* (2019) Structure, function and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase. Protein Science 28(3):478-486. IF = 2.400. DOI: 10.1002/pro.3571
Rubio-Cosials, A., Battistini, F., Gansen, A., Cuppari, A., Bernadó, P., Orozco, M., Langowski, J., Tóth, K * & M. Solà*. (2018) Protein flexibility and synergy of HMG domains underlie U-turn bending of DNA by TFAM in solution. Biophysical Journal 114 (10): 2386 – 2396. DOI: 10.1016/j.bpj.2017.11.3743
- Article mentioned in New and Notable (2017), DNA Bends the Knee to Transcription Factors, By G. Vámos & D. Rueda, Biophysical Journal, https://doi.org/10.1016/j.bpj.2017.10.047
Lyonnais, S*., Tarrés-Solé, A., Rubio-Cosials, A., Cuppari, A., Brito, R., Jaumot, J., Gargallo, R., Vilaseca, M., Silva, C., Granzhan, A., Teulade-Fichou, M.-P., Eritja, R. & Solà, M*. (2017) The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Scientific Reports, 7:43992. DOI: 10.1038/srep43992
Chakraborty, A., Lyonnais, S., Battistini, F., Hospital, A., Medici, G., Prohens, R., Orozco, M., Vilardell, J. & Solà, M*. (2017) DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p. Nucleic Acids Research, 45 (2): 951-967. DOI: 10.1093/nar/gkw1147. IF = 11.561.
Goulas, T., Mizgalska, D., Garcia-Ferrer, I., Kantyka, T., Guevara, T., Szmigielski, B., Sroka, A., Millan, C., Uson, I., Veillard, F., Potempa, B., Mydel, P., Solà, M.*, Potempa, J.* & Gomis-Ruth, F.X.* (2015). Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Scientific Reports 5: 11969. DOI: 10.1038/srep11969
Fernández-Millán, P., Lázaro, M., Cansiz-Arda, S., Gerhold, J. M., Rajala, N., Schmitz, C.A., Silva-Espiña, C., Gil, D., Bernadó, P., Valle, M.*, Spelbrink, J.N.* & Solà, M* (2015). The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Research 43: 4284-4295. DOI: 10.1093/nar/gkv189
Rubio-Cosials, A. & Solà, M.* (2013). U-turn DNA bending by human mitochondrial transcription factor A. Current Opinion of Structural Biology 23: 116-124. DOI: 10.1016/j.sbi.2012.12.004
Rubio-Cosials, A., Sydow, J.F., Jiménez-Menéndez, N., Fernández-Millán, P., Montoya, J., Jacobs, H.T., Coll, M., Bernadó, P. &Solà, M.* (2011). Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nature Structural and Molecular Biology 18: 1281–1289. DOI: 10.1038/nsmb.2160.
- Selected as “Article of the Month” in the December 2011 issue of the Journal of the Spanish Society of Biochemistry and Molecular Biology (SEBBM).
- First author (from the Structural MitoLab) was awarded the 2nd Prize (Accésit) “Fischer Scientific” to a Young Investigator at the XXXV SEBBM Meeting, held in Sevilla (Spain) in September 2012.
- Article mentioned in News and Views (Nature Structural and Molecular Biology 18: 1179–1181 (2011), DOI: 10.1038/nsmb.2167).
- Extraordinary PhD Award granted by the Doctorate School of the University of Barcelona.
Blanco, A.G., Canals, A., Bernués, J., Solà, M.& Coll, M. (2011). The structure of a transcription activation sub-complex reveals how σ70 is recruited to PhoB promoters. EMBO Journal 30: 3776 -3785. DOI: 10.1038/emboj.2011.271
Costenaro, L., Kaczmarska, Z., Arnan, C., Janowski, R., Coutard, B., Solà, M., Gorbalenya, A.E.; Norder, H.; Canard, B. & Coll, M. (2011). Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. Journal of Virology 85: 10764-73. DOI: 10.1128/JVI.05062-11
Nadal, M., Mas, P., Blanco, A.G., Arnan, C., Solà, M., Hart, D. & Coll, M. (2010). Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proceedings of the National Academy of Sciences USA 107: 16078-83. DOI: 10.1073/pnas.1007144107
Jiménez-Menéndez, N., Fernández-Millán, P., Rubio-Cosials, A., Arnan, C., Montoya, J., Jacobs, H.T., Bernadó, P., Coll, M., Usón, I. & Solà, M.* (2010). Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nature Structural and Molecular Biology 17: 891-3. DOI: 10.1038/nsmb.1859.
- Selected as “Article of the Month” in the issue August 2010 of the Journal of the Spanish Society of Biochemistry and Molecular Biology (SEBBM).
- First author (from the Structural MitoLab) was awarded the “José Tormo” Prize to a Young Investigator at the XXXIV SEBBM Meeting, held in Barcelona (Spain) in September 2011.
All publications
Spínola-Amilibia, M., Illanes-Vicioso, R., Ruiz-López, E., Colomer-Vidal, P., Ventura Rodriguez, F., Peces Pérez, R., Arias, C.F., Torroba, T., Solà, M.*, Arias-Palomo, E.*, Bertocchini, F.* (2023). Plastic degradation by insect hexamerins: Near-atomic resolution structures of the polyethylene-degrading proteins from the wax worm saliva. Science Advances 9, eadi6813, DOI: 10.1126/sciadv.adi6813
Tarrés-Solé, A., Battistini, F., Gerhold, J.M., Piétrement, O., Martínez-García, B., Ruiz-López, E., Lyonnais, S., Bernadó, P., Roca, J., Orozco, M., Le Cam, E., Sedman, J. & Solà, M.* (2023). Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism. Nucleic Acids Research, Volume 51 (11), Pages 5864–5882, doi.org/10.1093/nar/gkad397
Schmitz, C., Madej, M*., Nowakowska, Z., Cuppari A, et al., Potempa J *, Solà M* (2022). Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors. Nucleic Acids Research, Volume 50 (21), pp. 12558- 12577, doi.org/10.1093/nar/gkac1103
Sanluis-Verdes, A., Colomer-Vidal, P., Rodriguez-Ventura, F., Bello-Villarino, M., Spinola-Amilibia, M., Ruiz-Lopez, E., Illanes-Vicioso, R., Castroviejo, P., Aiese Cigliano, R., Montoya, M., Falabella, P., Pesquera, C., Gonzalez-Legarreta, L., Arias-Palomo, E., Solà, M., Torroba, T., Arias, C.F., Bertocchini, F. (2022) Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. Nature Communications 13, Article number: 5568. doi.org/10.1038/s41467-022-33127-w
Mizgalska, D., Goulas, T, Rodríguez-Banqueri, A., Veillarda, F., Madej, M., Małeckaa, E., Szczesniaka, K., Ksiazeka. M., Widziołeka, M., Guevara, T., Eckhardb, U., Solà, M.*, Potempa,J.*, Gomis-Rüth, F.-X.* (2021). Intermolecular latency regulates the essential C-terminal signal peptidase and sortase of the Porphyromonas gingivalis type-IX secretion system. PNAS 2021 Vol. 118 No. 40 e2103573118. DOI: 10.1073/pnas.2103573118
Piro-Mégy, C., Sarzi, E., Tarrés-Solé, A., Péquignot, M., Hensen, F., Quilès, M., Manes, G., Sénéchal, A., Bocquet, B., Cazevieille, C., Roubertie, A., Müller, A., Charif, M., Goudenège, D., Lenaers, G., Wilhelm, H., Kellner, U., Weisschuh, N., Wissinger, B., Zanlonghi, X., Spelbrink* J.N., Sola, M.*, Delettre, C.* (2020). Dominant mutations in SSBP1 impair mtDNA replication and cause optic atrophy and foveopathy. Journal of Clinical Investigation, 130(1):143-156. DOI: 10.1172/JCI128513
Cuppari, A., Fernández-Millán, P., Battistini, F., Tarrés-Solé, A., Lyonnais, S., Iruela, G., Ruiz-López, E., Enciso, Y., Rubio-Cosials, A., Prohens, R., Pons, M., Alfonso, C., Tóth, K., Rivas, G., Orozco, M., Solà, M. * (2019) DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region. Nucleic Acids Research, 47: 6519–6537. DOI: 10.1093/nar/gkz406
- Selected as “Article of the Month” in the July 2019 issue of the Journal of the Spanish Society of Biophysics (SEB), Biofísica.
Picchioni, D., Antolin-Fontes, A., Camacho, N., Schmitz, C., Pons-Pons, A., Rodriguez-Escribà, M., Machallekidou, Güller, M.N., Siatra, P., A., Carretero-Junquera, M., Serrano, A., Hovde, S.-L., Knobel, P.A., Novoa E., Solà-Vilarrubias, M., Kaguni, L.S., Stracker, T.H., Ribas de Pouplana, L. (2019) Mitochondrial Protein Synthesis and mtDNA Levels Coordinated through an Aminoacyl-tRNA Synthetase Subunit. Cell Reports, 271:40-47. DOI: 10.1016/j.celrep.2019.03.022
F. Hensen, A. Potter, S. Esvel, A. Tarrés-Solé, A. Chakraborty., M. Solà, J.N. Spelbrink* (2019) Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Research 47(7):3680-3698. DOI: 10.1093/nar/gkz047. IF= 11.147 (2018/2019).
Bereta, G., Goulas, T., Madej, M., Bielecka, E., Solà, M.*, Potempa, J.*, Gomis-Rüth, F.X.* (2019) Structure, function and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase. Protein Science 28(3):478-486. IF = 2.400. DOI: 10.1002/pro.3571
Rubio-Cosials, A., Battistini, F., Gansen, A., Cuppari, A., Bernadó, P., Orozco, M., Langowski, J., Tóth, K * & M. Solà*. (2018) Protein flexibility and synergy of HMG domains underlie U-turn bending of DNA by TFAM in solution. Biophysical Journal 114 (10): 2386 – 2396. DOI: 10.1016/j.bpj.2017.11.3743. IF = 3.665.
- Article mentioned in New and Notable (2017), DNA Bends the Knee to Transcription Factors, By G. Vámos & D. Rueda, Biophysical Journal, https://doi.org/10.1016/j.bpj.2017.10.047
Lyonnais, S*., Tarrés-Solé, A., Rubio-Cosials, A., Cuppari, A., Brito, R., Jaumot, J., Gargallo, R., Vilaseca, M., Silva, C., Granzhan, A., Teulade-Fichou, M.-P., Eritja, R. & Solà, M*. (2017) The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Scientific Reports, 7:43992. DOI: 10.1038/srep43992
Chakraborty, A., Lyonnais, S., Battistini, F., Hospital, A., Medici, G., Prohens, R., Orozco, M., Vilardell, J. & Solà, M*. (2017) DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p. Nucleic Acids Research, 45 (2): 951-967. DOI: 10.1093/nar/gkw1147. IF = 11.561.
Goulas, T., Mizgalska, D., Garcia-Ferrer, I., Kantyka, T., Guevara, T., Szmigielski, B., Sroka, A., Millan, C., Uson, I., Veillard, F., Potempa, B., Mydel, P., Solà, M.*, Potempa, J.* & Gomis-Ruth, F.X.* (2015). Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Scientific Reports 5: 11969. DOI: 10.1038/srep11969
Fernández-Millán, P., Lázaro, M., Cansiz-Arda, S., Gerhold, J. M., Rajala, N., Schmitz, C.A., Silva-Espiña, C., Gil, D., Bernadó, P., Valle, M.*, Spelbrink, J.N.* & Solà, M* (2015). The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Research 43: 4284-4295. DOI: 10.1093/nar/gkv189
Litwin, T., Solà, M., Holt, I., Neuman., K* (2015) A robust assay to measure DNA topology-dependent protein binding affinity. Nucleic Acids Research 43: e43 (Methods Online). DOI: 10.1093/nar/gku1381
Benabou, S., Ferreira, R., Aviñó, A., González, C., Lyonnais, S., Solà, M., Eritja, R., Jaumot, J., Gargallo, R. (2014). Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops. Biochimica et Biophysica Acta – General Subjects 1840: 41-52. DOI: 10.1016/j.bbagen.2013.08.028
Trillo-Muyo, S., Jasilionis, A., Domagalski, M.J., Chruszcz, M., Minor, W., Kuisiene, N., Arolas, J.L., Solà, M., Gomis-Rüth, F.X.* (2013). Ultratight crystal packing of a 10 kDa protein. Acta Crystallographica section D69: 464-470. DOI: 10.1107/S0907444912050135
Rubio-Cosials, A. & Solà, M.* (2013). U-turn DNA bending by human mitochondrial transcription factor A. Current Opinion of Structural Biology 23: 116-124. DOI: 10.1016/j.sbi.2012.12.004
Rubio-Cosials, A., Sydow, J.F., Jiménez-Menéndez, N., Fernández-Millán, P., Montoya, J., Jacobs, H.T., Coll, M., Bernadó, P. &Solà, M.* (2011). Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nature Structural and Molecular Biology 18: 1281–1289. DOI: 10.1038/nsmb.2160.
- Selected as “Article of the Month” in the December 2011 issue of the Journal of the Spanish Society of Biochemistry and Molecular Biology (SEBBM).
- First author (from the Structural MitoLab) was awarded the 2nd Prize (Accésit) “Fischer Scientific” to a Young Investigator at the XXXV SEBBM Meeting, held in Sevilla (Spain) in September 2012.
- Article mentioned in News and Views (Nature Structural and Molecular Biology 18: 1179–1181 (2011), DOI: 10.1038/nsmb.2167).
- Extraordinary PhD Award granted by the Doctorate School of the University of Barcelona.
Blanco, A.G., Canals, A., Bernués, J., Solà, M.& Coll, M. (2011). The structure of a transcription activation sub-complex reveals how σ70 is recruited to PhoB promoters. EMBO Journal 30: 3776 -3785. DOI: 10.1038/emboj.2011.271
Costenaro, L., Kaczmarska, Z., Arnan, C., Janowski, R., Coutard, B., Solà, M., Gorbalenya, A.E.; Norder, H.; Canard, B. & Coll, M. (2011). Structural basis for antiviral inhibition of the main protease, 3C, from human enterovirus 93. Journal of Virology 85: 10764-73. DOI: 10.1128/JVI.05062-11
Norder, H., De Palma, A.M., Selisko, B., Costenaro, L., Papageorgiou, N., Arnan, C., Coutard, B., Lantez, V., De Lamballerie, X., Baronti, C., Solà, M., Tan, J., Neyts, J., Canard, B., Coll, M., Gorbalenya, A.E. & Hilgenfeld, R. (2011). Picornavirus non-structural proteins as targets for new anti-virals with broad activity. Antiviral Research 89: 204-18. DOI: 10.1016/j.antiviral.2010.12.007
Nadal, M., Mas, P., Blanco, A.G., Arnan, C., Solà, M., Hart, D. & Coll, M. (2010). Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proceedings of the National Academy of Sciences USA 107: 16078-83. DOI: 10.1073/pnas.1007144107
Jiménez-Menéndez, N., Fernández-Millán, P., Rubio-Cosials, A., Arnan, C., Montoya, J., Jacobs, H.T., Bernadó, P., Coll, M., Usón, I. & Solà, M.* (2010). Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nature Structural and Molecular Biology 17: 891-3. DOI: 10.1038/nsmb.1859.
- Selected as “Article of the Month” in the issue August 2010 of the Journal of the Spanish Society of Biochemistry and Molecular Biology (SEBBM).
- First author (from the Structural MitoLab) was awarded the “José Tormo” Prize to a Young Investigator at the XXXIV SEBBM Meeting, held in Barcelona (Spain) in September 2011.
Monferrer, D., Tralau, T., Kertesz, M. A., Dix, I., Solà, M. & Usón, I. (2010). Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Molecular Microbiology 75: 1199-1214. DOI: 10.1111/j.1365-2958.2010.07043.x
Wójtowicz H, Guevara T, Tallant C, Olczak M, Sroka A, Potempa J, Solà M, Olczak T, Gomis-Rüth FX. (2009). Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathogens 5: e1000419. DOI: 10.1371/journal.ppat.1000419
Niemirowicz,G., Fernández, D., Solà, M., Cazzulo, J.-J., Avilés, F.X. & Gomis-Rüth, F.X. (2008). The molecular analysis of Trypanosoma cruzi metallocarboxypeptidase 1 provides insight into fold and substrate specificity. Molecular Microbiology75: 853–866. DOI: 10.1111/j.1365-2958.2008.06444.x
Sanglas, L., Valnickova, Z., Arolas, J. L., Pallarés, I., Guevara, T., Solà, M., Kristensen, T., Enghild, J.J., Avilés, F.X. & Gomis-Rüth, F.X. (2008). Structure of activated thrombin-activatable fibrinolysis inhibitor, a molecular link between coagulation and fibrinolysis. Molecular Cell 31: 598-606. DOI: 10.1016/j.molcel.2008.05.031
Mallorquí-Fernández, N., Manandhar, S.P., Mallorquí-Fernández, G., Usón, I., Wawrzonek, K., Kantyka, T., Solà, M., Thøgersen, I. B., Enghild, J., Potempa, J. & Gomis-Rüth, F.X. (2008). A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. Journal of Biological Chemistry 283: 2871-2882. DOI: 10.1074/jbc.M708481200
Tallant, C., García-Castellanos, R., Marrero, A., Canals, F., Yang, Y., Reymond, J.L., Solà, M., Baumann U. & Gomis-Rüth, F.X. (2007). Activity of ulilysin, an archaeal PAPP-A-related gelatinase and IGFBP-protease. Biological Chemistry 388: 1243-1253. DOI: 10.1515/BC.2007.143
Bayés, A., Fernandez, D., Solà, M., Marrero, A., Garcia-Piqué, S., Aviles, F.X., Vendrell, J. & Gomis-Rüth, F.X. (2007). Caught after the act: a human A-type metallocarboxypeptidase in a product complex with a cleaved hexapeptide. Biochemistry 46: 6921-6930. DOI: 10.1021/bi700480b
Saiyed, T., Paarman, I., Schmitt, B., Haeger, S., Solà, M., Schmalzing, G., Weissenhorn, W., &Betz, H. (2007). Molecular basis of gephyrin clustering at inhibitory synapses: role of G- and E domain interactions. Journal of Biological Chemistry 282: 5625-5632. DOI: 10.1074/jbc.M610290200. Q1; IF =
Arribas-Bosacoma, R., Kim, S.-K., Ferrer-Orta, C., Blanco, A.G., Pereira, P.J.B., Gomis-Rüth, F. X., Wanner, B.L., Coll, M. & Solà, M.* (2007). The X-ray crystal structures of two constitutively active mutants of the E. coli PhoB receiver domains gives insights into activation. Journal of Molecular Biology 366: 626-641. DOI: 10.1016/j.jmb.2006.11.038
García-Castellanos, R., Tallant, C., Solà, M., Baumann, U. & Gomis-Rüth, F.X. (2007). Substrate specificity of a metalloprotease of the pappalysin family revealed by an inhibitor and the product complex. Archives in Biochemistry and Biophysics 457: 57-72. DOI: 10.1016/j.abb.2006.10.004
Solà, M.*, Drew, D.L., Blanco, A.G., Gomis-Rüth, F. X.& Coll, M. (2006). The co-factor induced pre-active conformation in PhoB. Acta Crystallographica section D 62: 1046-1057. DOI: 10.1107/S0907444906024541
Guevara, T., Mallorquí-Fernández, N., García-Piqué, S., Petersen, G.E., Lauritzen, C., Pedersen, J., Arnau, J., Gomis-Rüth, F.X. & Solà, M.* (2006). Papaya glutamine cyclotransferase shows singular fivefold beta-propeller architecture that suggests a novel reaction mechanism. Biological Chemistry 387: 1479 – 1486. DOI: 10.1515/BC.2006.185
Gomis-Rüth, F.X., Solà, M., de la Cruz, F. & Coll, M. (2004). Coupling factors in macromolecular type-IV secretion machineries. Current Pharmaceutical Design 10: 1551-1565. DOI: 10.2174/1381612043384817
Solà, M., Bavro, V.N., Timmins, J., Franz, T., Ricard-Blum, S., Schoen, G., Ruigrok, R.W.H., Paarman, I., Saiyed, T., O’Sullivan, G., Schmitt, G., Betz, H. & Weissenhorn, W. (2004). Structural basis of dynamic glycine receptor clustering by gephyrin. EMBO Journal 23: 2510-2519. DOI: 10.1038/sj.emboj.7600256
Blanco, A.G., Sola, M., Gomis-Rüth, F.X. & Coll, M.(2002). Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure10: 61-73. DOI: 10.1016/S0969-2126(02)00761-X
Bavro, V.N., Sola, M., Bracher, A., Kneussel, M., Betz, H. & Weissenhorn, W. (2002). Crystal structure of the GABAA -receptor-associated protein GABARAP. EMBO Reports 3: 183-189. DOI: 10.1093/embo-reports/kvf026
Costa, M., Solà, M., del Solar, G., Eritja, R., Hernández-Arriaga, A.M., Espinosa, M., Gomis-Rüth, F.X. & Coll, M. (2001). Plasmid transcriptional repressor CopG oligomerises to render helical superstructures unbound and in complexes with oligonucleotides. Journal of Molecular Biology 310: 403-417. DOI: 10.1006/jmbi.2001.4760
Solà, M., Kneussel, M., Heck, I.S., Betz, H. & Weissenhorn, W. (2001) X-ray crystal structure of the trimeric N-terminal domain of Gephyrin. Journal of Biological Chemistry 276: 25294-25301. DOI: 10.1074/jbc.M101923200
Solà, M., López Hernández, E., Cronet, P., Lacroix, E., Serrano, L., Coll, M., & Párraga, A. (2000). Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. Journal of Molecular Biology 303: 213-225. DOI: 10.1006/jmbi.2000.4507
González, A., Pédélac, J.D., Solà, M., Gomis–Rüth, F.X., Coll, M., Samama, J.P. & Benini, S. (1999). Two-wavelength MAD phasing: in search of the optimal choice of wavelengths. Acta Crystallographica section D 55: 1449-1458. DOI: 10.1107/S0907444999006745
Solà, M., Gomis–Rüth, F.X., Serrano, L., González, A., & Coll, M. (1999). Three-dimensional crystal structure of the transcription factor PhoB receiver domain. Journal of Molecular Biology 85: 675-687. DOI: 10.1006/jmbi.1998.2326
Solà, M., Gomis–Rüth, F.X., Guasch, A., Serrano, L. & Coll, M. (1998). Overexpression, purification, crystallization and preliminary X-ray diffraction analysis of the receiver domain of PhoB. Acta Crystallographica section D54: 1460-1463. DOI: 10.1107/S0907444998006143
Gomis–Rüth, F.X., Solà, M., Pérez-Luque, R., Acebo, P., Alda, M. T., González, A., Espinosa, M., del Solar G. & Coll M. (1998). Overexpression, purification, crystallization and preliminary X-ray diffraction analysis of the pMV158-encoded plasmid transcriptional repressor protein CopG. FEBS letters 425: 161-165. DOI: 10.1016/S0014-5793(98)00219-1
Gomis–Rüth, F.X., Solà, M., Acebo, P., Párraga, A., González, A., Eritja, R., Espinosa, M., del Solar, G., & Coll, M. (1998). The structure of the plasmid-encoded transcriptional repressor CopG, unliganded and bound to its operator. EMBO Journal 17: 7404-7415. DOI: 10.1093/emboj/17.24.7404
Project Funding
Ongoing Projects
Reference: PID2021-129038NB-I00 (01.09.2022 – 2025)
Title: Análisis estructural de proteínas esenciales para la viabilidad mitocondrial y de patógenos-2
Proyecto PID2021-129038NB-I00 financiado por:

Concluded Projects
Reference: RTI2018-101015-B-100 (01.01.2019 – 31.08.2022)
Title: Structural analysis of proteins essential for mitochondria and pathogen viability
R&D project, Societal Challenges State Plan, Life Sciences Subplan, Ministry of Science, Innovation and Universities.
Proyecto RTI2018-101015-B-100 financiado por:

Reference: 2017 SGR 1192 (01.01.2017 – 31.12.2019)
Title: Computational methods and structure-function studies of proteins involved in mitochondrial pathologies and from pathogenic microorganisms (PATHOMET)
Consolidated Research Groups of Catalunya Grant, Generalitat of Catalunya.
Reference: BFU2015-70645-R
Title: Structural analysis of the mitocondrial genome and its regulation
R&Dproject, Fundamental Biology Plan, Molecular and Cellular Biology Subplan, Ministry of Economy and Competitiveness
Proyecto BFU2015-70645-R financiado por:

Reference: MDM-2014-0435
Title: Appointment of the Department of Structural Biology of IBMB as a “María de Maeztu” Unit of Excellence”
Total awarded: €2,000,000 (1.7.2015 – 31.12.2019)
Spanish Ministry of Economy and Competitivity.

Reference: FP7-HEALTH-2012-306029-2 “TRIGGER”
Title: King of hearts, joints and lungs; periodontal pathogens as etiologic factor in RA, CVD and COPD and their impact on treatment strategies
(1.4.2013 – 31.3.2017)
European Union STREP FP7 Project
Reference: FP7-PEOPLE-2011-290246 “RAPID”
Title: RAPID-Rheumatoid arthritis and periodontal inflammatory disease
Coordinator: Thomas Dietrich (Birmingham, UK). WP nº 3
(1.4.2012 – 31.3.2016)
European Union Marie Curie Initial Training Network (ITN) Project
Reference: BFU2012-33516/BMC
Title: Structural Basis of Mitochondrial Genome Regulation
(1.1.2013-31.12.2015)
R&Dproject, Fundamental Biology Plan, Molecular and Cellular Biology Subplan, Ministry of Economy and Competitiveness.
Reference: FP7-HEALTH-2010-261460
Title: Protein citrullination as a link between periodontal diseases and rheumatoid arthritis (RA) and target for development of novel drugs to treat RA
(1.11.2010 – 31.10.2014)
Coordinator: Peter Mydel (Göteborg, Sweden). WP nº 3
European Union STREP FP7 Project
Reference: BFU2009-07134/BMC
Title: Structural Biology of Mitochondrial Genome Regulation
(1.1.2010-31.12.2012)
Coordinator: Peter Mydel (Göteborg, Sweden). WP nº 3
R&Dproject, Fundamental Biology Plan, Molecular, Cellular and Genetics Biology Subplan, Ministry of Science and Innovation.
Reference: BFU2006-09593/BMC
R&D project, Fundamental Biology Plan, Molecular and Cellular Biology Subplan financed by the Ministry of Education and Science.
Reference: PIE 200820I045
C.S.I.C “Intramural” Project.
Vacancies/Jobs
Lab corner
No items available
Project gallery
No albums or photos uploaded yet.