New publication in eLife of the Jordan Lab in collaboration with the IBMB Imaging Platform showing that…
Condensin minimizes topoisomerase II-mediated entanglements of DNA in vivo
Inside the cell nucleus, the close juxtaposition of DNA segments, plus the DNA passage activity of topoisomerase II, lead to the formation of DNA knots and interlinks that jeopardize genome transactions. Recent studies conducted at the DNA Topology Lab of the IBMB-CSIC uncovered that some mechanism minimizes the knotting probability of intracellular DNA. Now, Dyson et al. have tested whether this minimization is achieved via the intrinsic capacity of topoisomerase II to simplify the equilibrium topology of DNA; or whether it is mediated by SMC complexes, whose capacity to extrude DNA loops can enforce the dissolution of DNA knots by topoisomerase II. They discovered that the low knotting probability of DNA does not rely on the simplification capacity of topoisomerase II, nor on the activities of cohesin or the smc5/6 complex. However, inactivation of condensin boosts the occurrence of DNA knots throughout the cell cycle. These findings suggest a role for the DNA loop extrusion activity of condensin in vivo and may explain why condensin disruption produces many alterations in interphase chromatin in addition to the persistence of sister chromatid interlinks in mitotic chromatin.
Reference:
Condensin minimizes topoisomerase II-mediated entanglements of DNA in vivo
Sílvia Dyson, Joana Segura, Belén Martínez-García, Antonio Valdés, and Joaquim Roca*
The EMBO Journal e105393 (2021)
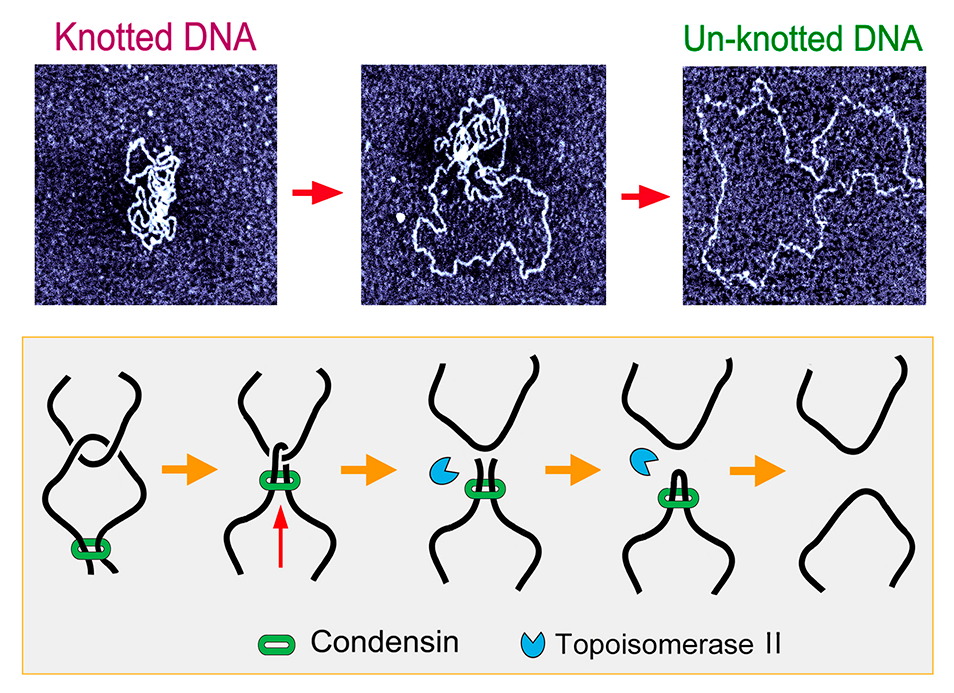
Top: progressive unknotting of a DNA molecule by means of topoisomerase II. Bottom: mechanistic model of minimization of DNA entanglements in vivo, in which the extruding activity of condensin tightens DNA crossovers and so enforces their dissolution by topoisomerase II.