This study provides important new insights into the regulation of alternative splicing in neurons by…
New publication in Nucleic Acids Research of the Jordan Lab showing that particular histone H1 variants (H1X and H1.4) accumulate at recently incorporated transposable elements in the human genome potentially participating in its repression
Abstract
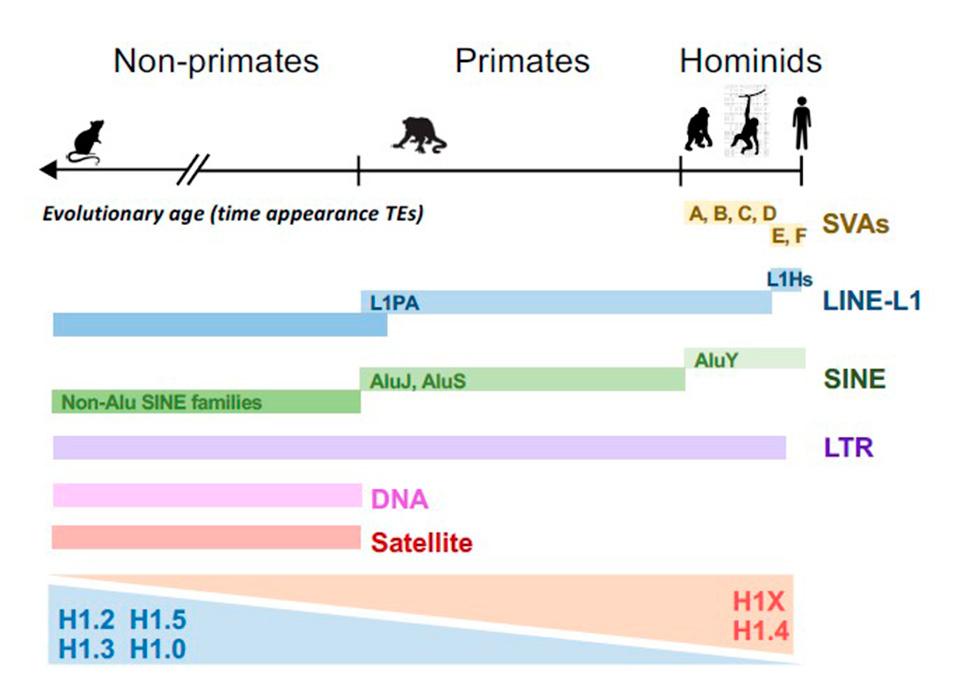
Histone H1, a vital component in chromatin structure, binds to linker DNA and regulates nuclear processes. We have investigated the distribution of histone H1 variants in a breast cancer cell line using ChIP-Seq. Two major groups of variants are identified: H1.2, H1.3, H1.5, and H1.0 are abundant in low GC regions (B compartment), while H1.4 and H1X preferentially localize in high GC regions (A compartment). Examining their abundance within transposable elements (TEs) reveals that H1X and H1.4 are enriched in recently-incorporated TEs (SVA and SINE-Alu), while H1.0/H1.2/H1.3/H1.5 are more abundant in older elements. Notably, H1X is particularly enriched in SVA families, while H1.4 shows the highest abundance in young AluY elements. Although low GC variants are generally enriched in LINE, LTR and DNA repeats, H1X and H1.4 are also abundant in a subset of recent LINE-L1 and LTR repeats. H1X enrichment at SVA and Alu is consistent across multiple cell lines. Further, H1X depletion leads to TE derepression, suggesting its role in maintaining TE repression. Overall, this study provides novel insights into the differential distribution of histone H1 variants among repetitive elements, highlighting the potential involvement of H1X in repressing TEs recently incorporated within the human genome.
Reference:
Mónica Salinas-Pena*, Núria Serna-Pujol*, Albert Jordan (2024) Genomic profiling of six human somatic histone H1 variants denotes that H1X accumulates at recently incorporated transposable elements. Nucleic Acids Research in press; doi: 10.1093/nar/gkae014
*these two authors contributed equally to this work