Lab presentation
We focus our research on the control of gene expression in human cells by chromatin organization, components and modifications, with a focus on the linker histone. The linker histone in mammals, participating in nucleosome spacing and higher-order chromatin structure, is a family of different histone H1 subtypes, including 7 somatic variants. It is poorly understood why so many H1 variants exist and whether they have particular features relevant to its functionality.
Knock-out experiments have suggested that the different variants are redundant and one isoform can substitute for another, but slowly new reports including ours are suggesting specific roles for the H1 variants, including involvement in specific gene expression regulation, and differential distribution in the human genome.
In a second line of research, we use an HIV promoter model to investigate the influence of chromatin organization at the integration site on HIV expression, with a focus on the role of heterochromatin on the establishment of viral latency, and we search for therapies for reactivation.
Member of the network Conexión Genoma del CSIC


Projects
FUNCTIONAL SPECIFICITY OF HUMAN HISTONE H1 VARIANTS AND ROLE IN CANCER
- Importance of H1 variants and the effect of their depletion on tumor progression and sensitivity to therapeutic interventions.
- Role and specificity of H1 variants in the organization of the genome and the nucleus.
- To study the functions of the H1 variants by identifying specific interactions with cellular factors.
The general objective of the present project is to understand why there are up to 7 variants of histone H1 in human somatic cells. Which functional differences exist between them, which occupation of the genome present, and which interactions establish with other nuclear factors or components of chromatin, and whether this depends on the cell type, cell stage, or is related to the cancerous process. The aim of the project is to understand how a structural component of chromatin is involved in the regulation of nuclear processes including the control of gene expression, with implications for proliferation, differentiation and deregulation of the tumor process.
We are an epigenomics laboratory where we use conventional molecular biology techniques to measure gene expression and protein accumulation, high-resolution confocal microscopy, proteomics, genomics (RNAseq, ChIPseq, ATACseq, HiC) and extensive bioinformatics. We use several human cancer cell models including 3D tumoroids/organoids and we plan to use single-cell analysis. We have developed all the tools to investigate the functionality of human H1 variants including specific antibodies and viral vectors for the expression or depletion of H1 variants.
Lab people

Albert Jordan
Principal investigator
After graduating in Biology at Autonomous University of Barcelona (UAB) Albert Jordan
went on to do his Ph.D. (1992-95) in the laboratory of Drs Jordi Barbé and Isidre Gibert
at the Dept. Genetics and Microbiology, UAB, where he was a Lecturer afterwards
(1996-98). In the meantime he spent five consecutive summer periods in Dr Peter
Reichard’s laboratory at the Karolinska Institute, Stockholm.
He then moved to San Francisco to undertake his postdoctoral work in the group of
Dr Eric Verdin at the Gladstone Institutes-UCSF (1999-2001), where he became interested
in the role of chromatin in human and viral gene expression control. In 2002 he got a
Ramon y Cajal appointment to join the group of Dr Miguel Beato at CRG (Barcelona) where he
became staff scientist afterwards. Later, in 2008, he got a permanent CSIC position to
start an independent group at IBMB, Dept. Molecular Genomics.
He has been Department director on 2011-13 and 2015-19. In parallel, Dr Jordan has been
appointed coordinator of the Molecular Biology section of the Catalan Society of
Biology (SCB) between 2009-16, coordinator of the Chromatin and Epigenetics section
since 2017, and member of the Directive Committee of SCB (Sections Coordinator,
Secretary) since 2012. Dr Jordan has been awarded several public and private grants,
directed 12 PhD thesis and published more than 50 papers.
Twitter ![]()
Coordinator of the group FUNCTIONAL SPECIFICITY AND REGULATION OF HISTONE H1-ESFUREH1 
Coordinator of the CHROMATIN and EPIGENETICS section of the CATALAN SOCIETY of BIOLOGY
https://scb.iec.cat/cromatina-i-epigenetica/

Carles Bonet

Núria Serna

Pau Homs

Ferran Fontdecaba
Past students
Daniel García
Mónica Salinas
Alberto Bustillos
Andrea Izquierdo
Erik Abner
Lluís Millan
Edurne Gallastegui
Albert Carcereny
Jaione Telleria Zufiaur
Mónica Salinas / personnel hired through aid for the recruitment of Novel research personnel (FI).
Selected publications
ORCID: 0000-0002-3970-8693 (Albert Jordan)
Salinas-Pena M*, Serna-Pujol N*, Jordan A (2024) Genomic profiling of six human somatic histone H1 variants denotes that H1X accumulates at recently incorporated transposable elements. Nucleic Acids Research Feb 28;52(4):1793-1813. doi: 10.1093/nar/gkae014
Salinas-Pena M, Rebollo E, Jordan A (2024) Imaging analysis of six human histone H1 variants reveals universal enrichment of H1.2, H1.3, and H1.5 at the nuclear periphery and nucleolar H1X presence. eLife Mar 26;12:RP91306. doi: 10.7554/eLife.91306
Pascal C, Zonszain J, Hameiri O, Tammer L, Ben-Salmon S, Roy VR, Eid M, Levy T, Rabe’a SA, Gargi C, Shalev N, Elbaz L, Hakim T, Lev-Maor G, Jordan A, Meshorer E, Ast G (2023) Human histone H1 variants impact splicing outcome by controlling RNA polymerase II elongation rate. Molecular Cell 83:3801-17. doi: 10.1016/j.molcel.2023.10.003
Fernández-Justel JM, Santa-María C, Martín-Vírgala S, Ramesh S, Ferrera-Lagoa A, Salinas-Pena M, Isoler-Alcaraz J, Maslon MM, Jordan A, Cáceres JF, Gómez M. (2022) Histone H1 regulates non-coding RNA turnover on chromatin in a m6A-dependent manner. Cell Reports. 40:111329. doi: 10.1016/j.celrep.2022.111329
Serna-Pujol N*, Salinas-Pena M*, Mugianesi F*, Le Dily F, Marti-Renom MA, Jordan A (2022) Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion. Nucleic Acids Research 05 April 2022. doi: 10.1093/nar/gkac226
Ponte I, Andrés M, Jordan A, Roque A (2021) Towards understanding the regulation of histone H1 somatic subtypes with OMICs. Journal of Molecular Biology 433: 166734. doi: 10.1016/j.jmb.2020.166734
Serna-Pujol N, Salinas-Pena M, Mugianesi F, Lopez-Anguita N, Torrent-Llagostera F, Izquierdo-Bouldstridge A, Marti-Renom MA, Jordan A (2021) TADs enriched in histone H1.2 strongly overlap with the B compartment, inaccessible chromatin and AT-rich Giemsa bands. The FEBS Journal 288: 1989-2013. doi: 10.1111/febs.15549
Abner E, Jordan A (2019) HIV ‘shock and kill’ therapy: in need of revision. Antiviral Research 166: 19-34. doi: 10.1016/j.antiviral.2019.03.008
Abner E, Stoszko M, Zeng L, Chen H-C, Izquierdo-Bouldstridge A, Konuma T, Zorita E, Fanunza E, Zhang Q, Mahmoudi T, Zhou M-M, Filion G, Jordan A (2018) A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for re-activation from other ‘shock’ drugs. Journal of Virology 2018 Apr 27;92(10). pii: e02056-17. doi: 10.1128/JVI.02056-17
Izquierdo-Bouldstridge A*, Bustillos A*, Bonet-Costa C, Aribau P, Garcia D, Dabad M, Esteve-Codina A, Pascual L, Peiro S, Esteller M, Murtha M, Millán-Ariño Ll, Jordan A (2017) Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Research 45(20): 11622-42. doi: 10.1093/nar/gkx746
Millán-Ariño Ll, Izquierdo-Bouldstridge A, Jordan A (2016) Specificities and genomic distribution of somatic mammalian histone H1 subtypes. BBA Gene Regulatory Mechanisms 1859(3): 510-19. doi: 10.1016/j.bbagrm.2015.10.013
Mayor R*, Izquierdo-Bouldstridge A*, Millán-Ariño Ll, Bustillos A, Sampaio C, Luque N, Jordan A (2015) Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. Journal of Biological Chemistry 290(12):7474-91. doi: 10.1074/jbc.M114.617324
Millán-Ariño Ll, Islam A, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, López-Bigas N, Jordan A (2014) Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Research 42:4474-4493. doi: 10.1093/nar/gku079
Gallastegui E, Marshall B, Vidal D, Sanchez-Duffhues G, Collado JA, Alvarez-Fernández C, Luque N, Terme JM, Gatell JM, Sánchez-Palomino S, Muñoz E, Mestres J, Verdin E, Jordan A (2012) Combination of biological screening in a cellular model of viral latency with virtual screening identifies novel compounds that reactivate HIV-1. Journal of Virology 86(7):3795-3808. doi: 10.1128/JVI.05972-11
Terme JM*, Sesé B*, Millán-Ariño L, Mayor R, Izpisua-Belmonte JC, Barrero MJ, Jordan A (2011) Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. Journal of Biological Chemistry 286(41):35347-57. doi: 10.1074/jbc.M111.281923
Gallastegui E, Millán-Zambrano G, Terme JM, Chávez S, Jordan A (2011) Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. Journal of Virology 85(7):3187-202. doi: 10.1128/JVI.01920-10
Sancho M, Diani E, Beato M, Jordan A (2008) Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLOS Genetics 2008 Oct;4(10):e1000227. doi: 10.1371/journal.pgen.1000227
Subtil-Rodríguez A, Millán-Ariño L, Quiles I, Ballaré C, Beato M, Jordan A (2008) Progesterone induction of 11ß-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and PR to a distal enhancer and polymerase tracking. Molecular and Cellular Biology 28: 3830-3849. doi: 10.1128/MCB.01217-07
Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, Jordan A, Beato M. (2006) Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Molecular Cell 24: 367-381. doi: 10.1016/j.molcel.2006.10.011
Larsson KM, Jordan A, Eliasson R, Reichard P, Logan DT, Nordlund P (2004) Structural mechanisms of allosteric substrate specificity regulation in a ribonucleotide reductase. Nature Structure and Molecular Biology 11: 1142-1149. doi: 10.1038/nsmb838
Jordan A, Bisgrove D, Verdin E. (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. The EMBO Journal 22: 1868-1877. doi: 10.1093/emboj/cdg188
Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivator. The EMBO Journal 20: 1726-1738. doi: 10.1093/emboj/20.7.1726
Jordan A, Reichard P. (1998) Ribonucleotide reductases. Annual Reviews of Biochemistry 67: 71-98. doi: 10.1146/annurev.biochem.67.1.71
Jordan A, Torrents E, Jeanthon C, Eliasson R, Hellman U, Wernstedt C, Barbé J, Gibert I, Reichard, P. (1997) B12-dependent ribonucleotide reductases from deeply rooted eubacteria are stucturally related to the aerobic enzyme from Escherichia coli. Proceedings of the National Academy of Sciences USA 94: 13487-13492. doi: 10.1073/pnas.94.25.13487
Jordan A, Pontis E, Atta M, Krrok M, Gibert I, Barbé J, Reichard P. (1994) A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proceedings of the National Academy of Sciences USA 91: 12892-12896. doi: 10.1073/pnas.91.26.12892
For a complete list of A. Jordan publications, search PubMed for:
(jordan a and roque a ) or (jordan a and salinas-pena M ) or (jordan a and serna-pujol n) or (jordan a and bonet-costa c) or (jordan a and izquierdo-bouldstridge a) or (jordan a and bartrons r) or (jordan a and terme jm) or (jordan a and chavez s) or (jordan a and beato m) or (jordan a and verdin e) or (jordan a and reichard p) or (jordan a and gibert i) or (jordan a and uhlin u) or (jordan a and eklund h) or (valles aj) or (jordan a and biochim biophys acta) or (jordan a and bachs o) or (jordan a and langley e) or (jordan a and szabo g) or (jordan a and abner e) or (jordan a and ast g)
All publications
ORCID: 0000-0002-3970-8693 (Albert Jordan)
Salinas-Pena M*, Serna-Pujol N*, Jordan A (2024) Genomic profiling of six human somatic histone H1 variants denotes that H1X accumulates at recently incorporated transposable elements. Nucleic Acids Research Feb 28;52(4):1793-1813. doi: 10.1093/nar/gkae014
Salinas-Pena M, Rebollo E, Jordan A (2024) Imaging analysis of six human histone H1 variants reveals universal enrichment of H1.2, H1.3, and H1.5 at the nuclear periphery and nucleolar H1X presence. eLife Mar 26;12:RP91306. doi: 10.7554/eLife.91306
Pascal C, Zonszain J, Hameiri O, Tammer L, Ben-Salmon S, Roy VR, Eid M, Levy T, Rabe’a SA, Gargi C, Shalev N, Elbaz L, Hakim T, Lev-Maor G, Jordan A, Meshorer E, Ast G (2023) Human histone H1 variants impact splicing outcome by controlling RNA polymerase II elongation rate. Molecular Cell 83:3801-17. doi: 10.1016/j.molcel.2023.10.003
Fernández-Justel JM, Santa-María C, Martín-Vírgala S, Ramesh S, Ferrera-Lagoa A, Salinas-Pena M, Isoler-Alcaraz J, Maslon MM, Jordan A, Cáceres JF, Gómez M. (2022) Histone H1 regulates non-coding RNA turnover on chromatin in a m6A-dependent manner. Cell Reports. 40:111329. doi: 10.1016/j.celrep.2022.111329
Imre L*, Firouzi Niaki E*, Bosire R, Nanasi Jr. P, Nagy P, Bacso Z, Hamidova N, Pommier Y, Jordan A, Szabo G (2022) Nucleosome destabilization by polyamines. Archives of Biochemistry and Biophysics. 05 April 2022. doi.org/10.1016/j.abb.2022.109184
Serna-Pujol N*, Salinas-Pena M*, Mugianesi F*, Le Dily F, Marti-Renom MA, Jordan A (2022) Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion. Nucleic Acids Research 05 April 2022. doi.org/10.1093/nar/
Ponte I, Andrés M, Jordan A, Roque A (2021) Towards understanding the regulation of histone H1 somatic subtypes with OMICs. Journal of Molecular Biology 433: 166734. doi: 10.1016/j.jmb.2020.166734
Serna-Pujol N, Salinas-Pena M, Mugianesi F, Lopez-Anguita N, Torrent-Llagostera F, Izquierdo-Bouldstridge A, Marti-Renom MA, Jordan A (2021) TADs enriched in histone H1.2 strongly overlap with the B compartment, inaccessible chromatin and AT-rich Giemsa bands. The FEBS Journal 288: 1989-2013. doi: 10.1111/febs.15549
Abner E, Jordan A (2019) HIV ‘shock and kill’ therapy: in need of revision. Antiviral Research 166: 19-34. doi: 10.1016/j.antiviral.2019.03.008
Salmerón-Hernández A, Noriega-Reyes MY, Jordan A, Baranda-Avila N, Langley E (2019) BCAS2 Enhances Carcinogenic Effects of Estrogen Receptor Alpha in Breast Cancer Cells. International Journal of Molecular Sciences Feb 22;20(4). pii: E966. doi: 10.3390/ijms20040966
Carbonell A, Fueyo R, Izquierdo-Bouldstridge A, Moreta C, Jordan A (2018) Epigenetic mechanisms in health and disease: BCEC 2017. Epigenetics 13(3): 331-341. doi: 10.1080/15592294.2018.1434391
Abner E, Stoszko M, Zeng L, Chen H-C, Izquierdo-Bouldstridge A, Konuma T, Zorita E, Fanunza E, Zhang Q, Mahmoudi T, Zhou M-M, Filion G, Jordan A (2018) A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for re-activation from other ‘shock’ drugs. Journal of Virology 2018 Apr 27;92(10). pii: e02056-17. doi: 10.1128/jvi.02056-17
Izquierdo-Bouldstridge A*, Bustillos A*, Bonet-Costa C, Aribau P, Garcia D, Dabad M, Esteve-Codina A, Pascual L, Peiro S, Esteller M, Murtha M, Millán-Ariño Ll, Jordan A (2017) Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Research 45(20): 11622-42. doi: 10.1093/nar/gkx746
Perearnau A, Orlando S, Islam A, Gallastegui E, Martínez J, Jordan A, Bigas A, Aligué R, Pujol MJ, Bachs O (2017) p27Kip1, PCAF and PAX5 cooperate in the transcriptional regulation of specific target genes. Nucleic Acids Research 45(9):5086-99. doi: 10.1093/nar/gkx075
Castaño J, Morera C, Sesé B, Boue S, Bonet-Costa C, Marti M, Roque A, Jordan A, Barrero MJ (2016) SETD7 regulates the differentiation of human embryonic stem cells. PLOS One doi: 10.1371/journal.pone.0149502.
Jordan A (2016) Editorial-Histone H1 in gene expression and development. BBA Gene Regulatory Mechanisms 1859(3): 429-30. doi: 10.1016/j.bbagrm.2016.01.001
Millán-Ariño Ll, Izquierdo-Bouldstridge A, Jordan A (2016) Specificities and genomic distribution of somatic mammalian histone H1 subtypes. BBA Gene Regulatory Mechanisms 1859(3): 510-19. doi: 10.1016/j.bbagrm.2015.10.013
Noriega-Reyes MY, Rivas-Torres MA, Oñate-Ocaña LF, Jordan-Vallés A, Baranda-Avila N, Langley E (2015) Novel role for PINX1 as a coregulator of nuclear hormone receptors. Molecular and Cellular Endocrinology 414: 9-18. doi: 10.1016/j.mce.2015.07.011
Mayor R*, Izquierdo-Bouldstridge A*, Millán-Ariño Ll, Bustillos A, Sampaio C, Luque N, Jordan A (2015) Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. Journal of Biological Chemistry 290(12):7474-91. doi: 10.1074/jbc.M114.617324
Terme JM, Millán-Ariño Ll, Mayor R, Luque N, Izquierdo-Bouldstridge A, Bustillos A, Sampaio C, Canes J, Font I, Sima N, Sancho M, Torrente L, Forcales S, Roque A, Suau P, Jordan A (2014) Dynamics and dispensability of variant-specific histone H1 Lys-26/Ser-27 and Thr-165 post-translational modifications. FEBS Letters 588(14):2353-62. doi: 10.1016/j.febslet.2014.05.035
Millán-Ariño Ll, Islam A, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, López-Bigas N, Jordan A (2014) Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Research 42:4474-4493. doi: 10.1093/nar/gku079
Gallastegui E, Marshall B, Vidal D, Sanchez-Duffhues G, Collado JA, Alvarez-Fernández C, Luque N, Terme JM, Gatell JM, Sánchez-Palomino S, Muñoz E, Mestres J, Verdin E, Jordan A (2012) Combination of biological screening in a cellular model of viral latency with virtual screening identifies novel compounds that reactivate HIV-1. Journal of Virology 86(7):3795-3808. doi: 10.1128/JVI.05972-11
Novellasdemunt L, Obach M, Millán-Ariño Ll, Manzano A, Ventura F, Rosa JL, Jordan A, Navarro-Sabate A, Bartrons R (2012) Progestins activate 6-Phosphofructo-2-kinase/Fructose-2,6-Bisphosphatase (PFKFB3) in breast cancer cells. Biochemical Journal 442(2):345-56. doi: 10.1042/bj20111418
Terme JM*, Sesé B*, Millán-Ariño L, Mayor R, Izpisua-Belmonte JC, Barrero MJ, Jordan A (2011) Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. Journal of Biological Chemistry 286(41):35347-57. doi: 10.1074/jbc.M111.281923
Gallastegui E, Millán-Zambrano G, Terme JM, Chávez S, Jordan A (2011) Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. Journal of Virology 85(7):3187-202. doi: 10.1128/JVI.01920-10
Terme JM, Calvignac S, Duc Dodon M, Gazzolo L, Jordan A (2009) E box motifs as mediators of proviral latency of human retroviruses. Retrovirology 6: 81. doi: 10.1186/1742-4690-6-81
Quiles I, Millán-Ariño L, Subtil-Rodríguez A, Miñana B, Spinedi N, Ballaré C, Beato M, Jordan A (2009) Mutational analysis of progesterone receptor functional domains in stable cell lines delineates sets of genes regulated by different mechanisms. Molecular Endocrinology 23: 809-26. doi: 10.1210/me.2008-0454
Vanti M*, Gallastegui E*, Respaldiza I, Rodríguez-Gil A, Gómez-Herreros F, Jimeno-González S, Jordan A and Chávez S (2009) Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLOS Genetics 2009 Jan;5(1):e1000339. doi: 10.1371/journal.pgen.1000339
Sancho M, Diani E, Beato M, Jordan A (2008) Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLOS Genetics 2008 Oct;4(10):e1000227. doi: 10.1371/journal.pgen.1000227
Subtil-Rodríguez A, Millán-Ariño L, Quiles I, Ballaré C, Beato M, Jordan A (2008) Progesterone induction of 11ß-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and PR to a distal enhancer and polymerase tracking. Molecular and Cellular Biology 28: 3830-3849. doi: 10.1128/MCB.01217-07
Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, Jordan A, Beato M (2008) Convergence on chromatin of non-genomic and genomic pathways of hormone signaling. Journal of Steroid Biochemistry and Molecular Biology 109: 344-349. doi: 10.1016/j.jsbmb.2008.03.015
Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, Jordan A, Beato M. (2006) Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Molecular Cell 24: 367-381. doi: 10.1016/j.molcel.2006.10.011
Larsson KM, Jordan A, Eliasson R, Reichard P, Logan DT, Nordlund P (2004) Structural mechanisms of allosteric substrate specificity regulation in a ribonucleotide reductase. Nature Structure and Molecular Biology 11: 1142-1149. doi: 10.1038/nsmb838
Jordan A, Bisgrove D, Verdin E. (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. The EMBO Journal 22: 1868-1877. doi: 10.1093/emboj/cdg188
Pion M, Jordan A, Biancotto A, Dequiedt F, Gondois-Rey F, Rondeau S, Vigne R, Hejnar J, Verdin E, Hirsch I (2003) Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. Journal of Virology 77: 4025-4032. doi: 10.1128/jvi.77.7.4025-4032.2003
Jordan A, Defechereux P, Verdin E (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivator. The EMBO Journal 20: 1726-1738. doi: 10.1093/emboj/20.7.1726
Jordan A, Torrents E, Sala I, Hellman U, Gibert I, Reichard, P (1999) Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. Journal of Bacteriology 181: 3974-3980. doi: 10.1128/2Fjb.181.13.3974-3980.1999
Eliasson R, Pontis E, Jordan A, Reichard P (1999) Allosteric control of three B12-dependent (class II) ribonucleotide reductases. Implication for the evolution of ribonucleotide reduction. Journal of Biological Chemistry 274: 7182-7189. doi: 10.1074/jbc.274.11.7182
Jordan A, Reichard P. (1998) Ribonucleotide reductases. Annual Reviews of Biochemistry 67: 71-98. doi: 10.1146/annurev.biochem.67.1.71
Fieschi F, Torrents E, Toulokhonova L, Jordan A, Hellman U, Gibert I, Barbé J, Karlsson M, Sjöberg B-M. (1998) The manganese-containing ribonucleotide reductase of Corynebacterium ammoniagenes is a class Ib enzyme. Journal of Biological Chemistry 273: 4329-4337. doi: 10.1074/jbc.273.8.4329
Jordan A, Torrents E, Jeanthon C, Eliasson R, Hellman U, Wernstedt C, Barbé J, Gibert I, Reichard, P. (1997) B12-dependent ribonucleotide reductases from deeply rooted eubacteria are stucturally related to the aerobic enzyme from Escherichia coli. Proceedings of the National Academy of Sciences USA 94: 13487-13492. doi: 10.1073/pnas.94.25.13487
Jordan A, Åslund F, Pontis E, Reichard P, Holmgren A. (1997) Characterization of E. coli NrdH: a glutaredoxin-like protein with thioredoxin-like activity profile. Journal of Biological Chemistry 2: 18044-18050. doi: 10.1074/jbc.272.29.18044
Eliasson R, Pontis E, Jordan A, Reichard P. (1996) Allosteric regulation of the third ribonucleotide reductase (NrdEF enzyme) from Enterobacteriaceae. Journal of Biological Chemistry 271: 26582-26587. doi: 10.1074/jbc.271.43.26582
Jordan A, Pontis E, Åslund F, Hellman U, Gibert I, Reichard P. (1996) The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF-enzyme and a new electron transport protein. Journal of Biological Chemistry 27: 8779-8785. doi: 10.1074/jbc.271.15.8779
Jordan A, Aragall E, Gibert I, Barbé J. (1996) Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Molecular Microbiology 19:777-790. doi: 10.1046/j.1365-2958.1996.424950.x
Jordan A, Pontis E, Atta M, Krrok M, Gibert I, Barbé J, Reichard P. (1994) A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proceedings of the National Academy of Sciences USA 91: 12892-12896. doi: 10.1073/pnas.91.26.12892
Jordan A, Gibert I, Barbé J. (1994) Cloning and sequencing of the genes from S. typhimurium encoding a new bacterial ribonucleotide reductase. Journal of Bacteriology 176: 3420-3427. doi: 10.1128/jb.176.11.3420-3427.1994
For a complete list of A. Jordan publications, search PubMed for:
(jordan a and roque a ) or (jordan a and salinas-pena M ) or (jordan a and serna-pujol n) or (jordan a and bonet-costa c) or (jordan a and izquierdo-bouldstridge a) or (jordan a and bartrons r) or (jordan a and terme jm) or (jordan a and chavez s) or (jordan a and beato m) or (jordan a and verdin e) or (jordan a and reichard p) or (jordan a and gibert i) or (jordan a and uhlin u) or (jordan a and eklund h) or (valles aj) or (jordan a and biochim biophys acta) or (jordan a and bachs o) or (jordan a and langley e) or (jordan a and szabo g) or (jordan a and abner e) or (jordan a and ast g)

Project funding
Functional specificity of human histone H1 variants and role in cancer.
- AGENCY: Ministerio de Ciencia e Innovación – Plan Nacional BFU
- PERIOD: 2021 – 2024.
- IP: Albert Jordan Vallès
Proyecto PID2020-112783GB-C21 financiado por:

Especificidad funcional de las variantes de la histona H1 humana:
- AGENCY: Ministerio de Ciencia e Innovación – Plan Nacional BFU
- PERIOD: 2018 – 2020
- IP: Albert Jordan Vallès
Proyecto BFU2017-82805-C2-1-P financiado por:

Especificidad funcional de las variantes de la histona H1 humana.
- AGENCY: Ministerio de Ciencia e Innovación – Plan Nacional BFU
- PERIOD: 2015 – 2017
- IP: Albert Jordan Vallès
Especificidad funcional de las variantes de la histona H1 humanas:
- AGENCY: Ministerio de Ciencia e Innovación – Plan Nacional BFU
- PERIOD: 2012 – 2014
- IP: Albert Jordan Vallès
Determinantes genómicos del estado latente del VIH-1 y estrategias terapéuticas:
- AGENCY: FIPSE
- PERIOD: 2011 – 2013
- IP: Albert Jordan Vallès
Estrategias terapéuticas para la reactivación del Virus de la Immunodeficiencia Humana en células latentes:
- AGENCY: Fundación Eugenio Rodríguez Pascual
- PERIOD: 2011
- IP: Albert Jordan Vallès
Genómica estructural y aplicada:
- AGENCY: AGAUR-DURSI – Grupos reconocidos
- PERIOD: 2009 – 2013
- IP: Joaquim Roca / Marian Martínez Balbas / Xavier de la Cruz / Albert Jordan
Especificidad de las variantes de la histona H1 humanas en la estructura de la cromatina, expresión génica y proliferación:
- AGENCY: Ministerio de Ciencia e Innovación – Plan Nacional BFU
- PERIOD: 2009 – 2011
- IP: Albert Jordan Vallès
Reactivación del VIH en células T latentemente infectadas in vitro e in vivo:
- AGENCY: FIPSE
- PERIOD: 2006 – 2009
- IP: Albert Jordan Vallès
Latencia del VIH. Estudio del mecanismo molecular y búsqueda de dianas para la reactivación:
- AGENCY Instituto de Salud Carlos III – Fondo de Investigación Sanitaria
- PERIOD: 2006 – 2008
- IP: Albert Jordan Vallès
Regulación transcripcional y reorganización de la cromatina en los promotores pS2, 11 beta-HSD y MMTV en células de cáncer de mama en respuesta a hormonas esteroideas:
- AGENCY: Ministerio de Ciencia y Tecnología – Plan Nacional SAF
- PERIOD: 2002 – 2005
- IP: Albert Jordan Vallès
Vacancies/Jobs
Candidates, send CV and an introductory letter (experience, references, motivation…) by email to albert.jordan@ibmb.csic.es
Pre- and post-doc candidates Chromatin regulation of human and viral gene expression
Pre- and post-doc candidates with an outstanding CV to apply to competitive fellowships will be always…
Lab corner
IMAGING ANALYSIS OF SIX HUMAN HISTONE H1 VARIANTS REVEALS UNIVERSAL ENRICHMENT OF H1.2, H1.3, AND H1.5 AT THE NUCLEAR PERIPHERY AND NUCLEOLAR H1X PRESENCE
Histone H1 participates in chromatin condensation and regulates nuclear processes. Human somatic cells may contain up to seven histone H1 variants, although their functional heterogeneity is not fully understood. Here, we have profiled the differential nuclear distribution of the somatic H1 repertoire in human cells through imaging techniques including super-resolution microscopy. H1 variants exhibit characteristic distribution patterns in both interphase and mitosis. H1.2, H1.3, and H1.5 are universally enriched at the nuclear periphery in all cell lines analyzed and co-localize with compacted DNA. H1.0 shows a less pronounced peripheral localization, with apparent variability among different cell lines. On the other hand, H1.4 and H1X are distributed throughout the nucleus, being H1X universally enriched in high-GC regions and abundant in the nucleoli. Interestingly, H1.4 and H1.0 show a more peripheral distribution in cell lines lacking H1.3 and H1.5. The differential distribution patterns of H1 suggest specific functionalities in organizing lamina-associated domains or nucleolar activity, which is further supported by a distinct response of H1X or phosphorylated H1.4 to the inhibition of rDNA transcription. Moreover, H1 variants depletion affects chromatin structure in a variant-specific manner. Concretely, H1.2 knock-down, either alone or combined, triggers a global chromatin decompaction. Overall, imaging has allowed us to distinguish H1 variants distribution beyond the segregation in two groups denoted by previous ChIP-seq determinations. Our results support H1 variants heterogeneity and suggest that variant-specific functionality can be shared between different cell types.
Salinas-Pena M, Rebollo E, Jordan A (2024) Imaging analysis of six human histone H1 variants reveals universal enrichment of H1.2, H1.3, and H1.5 at the nuclear periphery and nucleolar H1X presence. eLife Mar 26;12:RP91306. doi: 10.7554/eLife.91306.1
GENOMIC PROFILING OF SIX HUMAN SOMATIC HISTONE H1 VARIANTS DENOTES THAT H1X ACCUMULATES AT RECENTLY INCORPORATED TRANSPOSABLE ELEMENTS
Histone H1, a vital component in chromatin structure, binds to linker DNA and regulates nuclear processes. We have investigated the distribution of histone H1 variants in a breast cancer cell line using ChIP-Seq. Two major groups of variants are identified: H1.2, H1.3, H1.5, and H1.0 are abundant in low GC regions (B compartment), while H1.4 and H1X preferentially localize in high GC regions (A compartment). Examining their abundance within transposable elements (TEs) reveals that H1X and H1.4 are enriched in recently-incorporated TEs (SVA and SINE-Alu), while H1.0/H1.2/H1.3/H1.5 are more abundant in older elements. Notably, H1X is particularly enriched in SVA families, while H1.4 shows the highest abundance in young AluY elements. Although low GC variants are generally enriched in LINE, LTR and DNA repeats, H1X and H1.4 are also abundant in a subset of recent LINE-L1 and LTR repeats. H1X enrichment at SVA and Alu is consistent across multiple cell lines. Further, H1X depletion leads to TE derepression, suggesting its role in maintaining TE repression. Overall, this study provides novel insights into the differential distribution of histone H1 variants among repetitive elements, highlighting the potential involvement of H1X in repressing TEs recently incorporated within the human genome.
Salinas-Pena M*, Serna-Pujol N*, Jordan A (2024) Genomic profiling of six human somatic histone H1 variants denotes that H1X accumulates at recently incorporated transposable elements. Nucleic Acids Research Feb 28;52(4):1793-1813. doi: 10.1093/nar/gkae014
COORDINATED CHANGES IN GENE EXPRESSION, H1 VARIANT DISTRIBUTION AND GENOME 3D CONFORMATION IN RESPONSE TO H1 DEPLETION
Researchers of the IBMB have investigated for the first time the genomic distribution of five histone H1 variants in a human cancer cell line and the consequences of H1 depletion on the 3D conformation of the genome, which will help to better understand the function of this structural component of chromatin. Up to seven members of the histone H1 family may contribute to chromatin compaction and its regulation in human somatic cells. In breast cancer cells, knock-down of multiple H1 variants deregulates many genes, promotes the appearance of genome-wide accessibility sites and triggers an interferon response via activation of heterochromatic repeats. However, how these changes in the expression profile relate to the re-distribution of H1 variants as well as to genome conformational changes have not been yet studied. Here, we combined ChIP-seq of five endogenous H1 variants with Chromosome Conformation Capture analysis in wild-type and H1.2/H1.4 knock-down T47D cells. The results indicate that H1 variants coexist in the genome in two large groups depending on the local GC content and that their distribution is robust with respect to H1 depletion. Despite the small changes in H1 variants distribution, knock-down of H1 translated into more isolated but de-compacted chromatin structures at the scale of topologically associating domains (TADs). Such changes in TAD structure correlated with a coordinated gene expression response of their resident genes. This is the first report describing simultaneous profiling of five endogenous H1 variants and giving functional evidence of genome topology alterations upon H1 depletion in human cancer cells.
Serna-Pujol N*, Salinas-Pena M*, Mugianesi F*, Le Dily F, Marti-Renom MA, Jordan A (2022) Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion. Nucleic Acids Research 05 April 2022. https://doi.org/10.1093/nar/gkac226
TADS ENRICHED IN HISTONE H1.2 STRONGLY OVERLAP WITH THE B COMPARTMENT, INACCESSIBLE CHROMATIN AND AT-RICH GIEMSA BANDS
An epigenetic characterization of Giemsa bands (G bands) performed by researchers of the IBMB has revealed their utility as epigenetic units to investigate the differential distribution of linker histones. Giemsa staining of metaphase chromosomes results in a characteristic banding useful for identification of chromosomes and its alterations. Staining of G positive bands decreases with GC content. High GC bands are enriched in active histone marks, RNA polymerase II and SINEs, and associate to gene richness, gene expression and early replication. Histone H1 variants also distribute heterogeneously among G bands: H1X is enriched at high GC bands and H1.2 is abundant at low GC, compacted bands. According to epigenetic features and H1 content, G bands can be organized in clusters useful to compartmentalize the genome. Besides, Hi-C analysis has shown that TADs with high H1.2/H1X ratio strongly overlap with B compartment, late replicating and inaccessible chromatin, and low GC bands. Serna et al. propose that GC content is a strong driver of chromatin compaction and 3D genome organization, that Giemsa staining recapitulates this organization denoted by high-throughput techniques, and that H1 variants distribute at distinct chromatin domains.
Serna-Pujol N, Salinas-Pena M, Mugianesi F, Lopez-Anguita N, Torrent-Llagostera F, Izquierdo-Bouldstridge A, Marti-Renom MA, Jordan A (2020) TADs enriched in histone H1.2 strongly overlap with the B compartment, inaccessible chromatin and AT-rich Giemsa bands. FEBS Journal doi: 10.1111/febs.15549.
HIV ‘SHOCK AND KILL’ THERAPY: IN NEED OF REVISION
The implementation of antiretroviral therapy 23 years ago has rendered HIV infection clinically manageable. However, the disease remains incurable, since it establishes latent proviral reservoirs, which in turn can stochastically begin reproducing viral particles throughout the patient’s lifetime. Viral latency itself depends in large part on the silencing environment of the infected host cell, which can be chemically manipulated. “Shock and kill” therapy intends to reverse proviral quiescence by inducing transcription with pharmaceuticals and allowing a combination of antiretroviral therapy, host immune clearance and HIV-cytolysis to remove latently infected cells, leading to a complete cure. Over 160 compounds functioning as latency-reversing agents (LRAs) have been identified to date, but none of the candidates has yet led to a promising functional cure. In this review we describe the variety of existing classes of LRAs, discuss their current drawbacks and highlight the potential for combinatorial “shocktail” therapies for potent proviral reactivation.
Abner E, Jordan A (2019) HIV ‘shock and kill’ therapy: in need of revision. Antiviral Research 166: 19-34. doi: 10.1016/j.antiviral.2019.03.008.
A NEW QUINOLINE BRD4 INHIBITOR TARGETS A DISTINCT LATENT HIV-1 RESERVOIR FOR RE- ACTIVATION FROM OTHER ‘SHOCK’ DRUGS
Researchers of the IBMB have shown that a previously identified Human Immunodeficiency Virus (HIV) latency-reversing agent (MMQO) acts as a bromodomain inhibitor. Despite of very different chemical structures, MMQO mimics the action of JQ1, a well-known bromodomain inhibitor, and binds to the bromodomain and extraterminal domain (BET) family protein BRD4. Utilizing barcoded HIV-1minigenomes, we demonstrate that PKC pathway activators, HDAC and bromodomain inhibitors all target different subsets of proviral integrations. Considering the fundamental differences of these compounds and the synergies displayed between them, we propose that the field should concentrate on investigating the development of combinatory “shock” cocktail therapies for an improved reservoir reactivation aiming to eradicate the latent functional proportion of HIV-1 proviruses in a patient.
Abner E, Stoszko M, Zeng L, Chen H-C, Izquierdo-Bouldstridge A, Konuma T, Zorita E, Fanunza E, Zhang Q, Mahmoudi T, Zhou M-M, Filion G, Jordan A (2018) A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for re-activation from other ‘shock’ drugs. Journal of Virology 92(10). pii: e02056-17. doi: 10.1128/JVI.02056-17
HISTONE H1 DEPLETION TRIGGERS AN INTERFERON RESPONSE IN CANCER CELLS VIA ACTIVATION OF HETEROCHROMATIC REPEATS
Researchers of the IBMB have shown that simultaneous depletion of particular histone H1 variants in human breast cancer cells triggers the cellular interferon response, originally designed to fight against invading pathogen-associated nucleic acids such as DNA or RNA viruses. They have shown that this is due to the derepression of heterochromatic non-coding RNAs such as satellites and endogenous retroviruses, which may be naturally repressed with the participation of particular histone H1 subtypes such as H1.2 and H1.4.
Izquierdo-Bouldstridge A*, Bustillos A*, Bonet-Costa C, Aribau P, Garcia D, Dabad M, Esteve-Codina A, Pascual L, Peiro S, Esteller M, Murtha M, Millán-Ariño Ll, Jordan A (2017) Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Research 45(20):11622-42. doi: 10.1093/nar/gkx746.
Project gallery
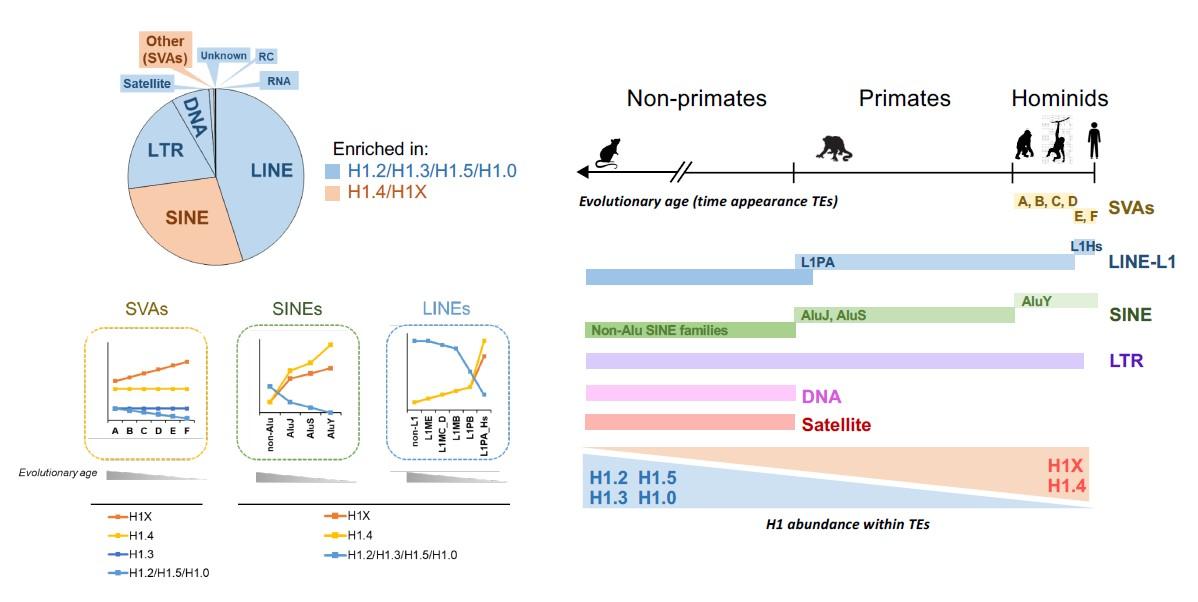
GENOMIC PROFILING OF SIX HUMAN SOMATIC HISTONE H1 VARIANTS DENOTES THAT H1X ACCUMULATES AT RECENTLY INCORPORATED TRANSPOSABLE ELEMENTS (Nucleic Acids Research 2024)
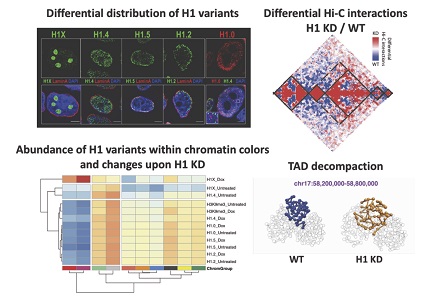
COORDINATED CHANGES IN GENE EXPRESSION, H1 VARIANT DISTRIBUTION AND GENOME 3D CONFORMATION IN RESPONSE TO H1 DEPLETION (Nucleic Acids Research 2022)
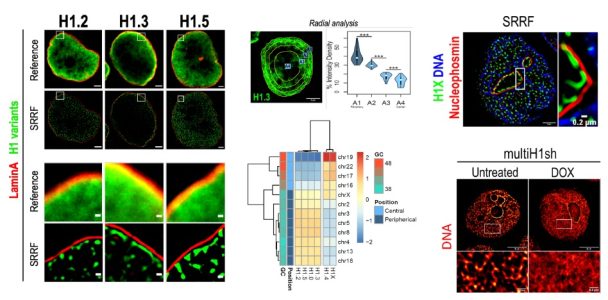
IMAGING ANALYSIS OF SIX HUMAN HISTONE H1 VARIANTS REVEALS UNIVERSAL ENRICHMENT OF H1.2, H1.3, AND H1.5 AT THE NUCLEAR PERIPHERY AND NUCLEOLAR H1X PRESENCE (eLife 2023)
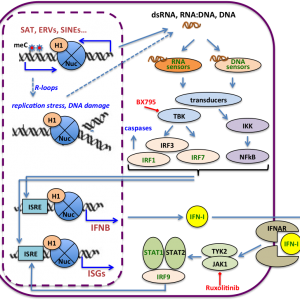
HISTONE H1 DEPLETION TRIGGERS AN INTERFERON RESPONSE IN CANCER CELLS VIA ACTIVATION OF HETEROCHROMATIC REPEATS (Nucleic Acids Research 2017)
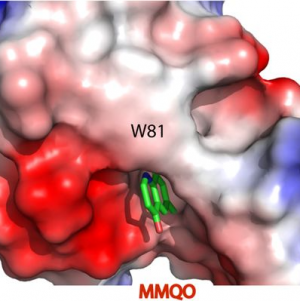
A NEW QUINOLINE BRD4 INHIBITOR TARGETS A DISTINCT LATENT HIV-1 RESERVOIR FOR REACTIVATION FROM OTHER ‘SHOCK’ DRUGS (Journal of Virology 2018)
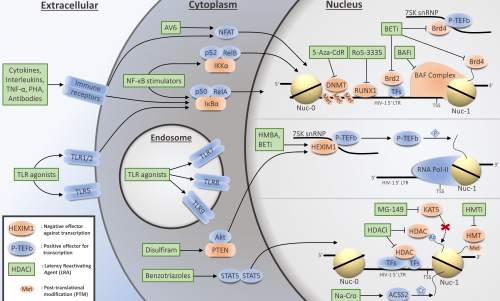
HIV ‘SHOCK AND KILL’ THERAPY: IN NEED OF REVISION (Antiviral Research 2019)
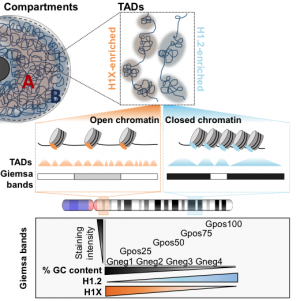
TADS ENRICHED IN HISTONE H1.2 STRONGLY OVERLAP WITH THE B COMPARTMENT, INACCESSIBLE CHROMATIN AND AT-RICH GIEMSA BANDS (FEBS Journal 2020)


