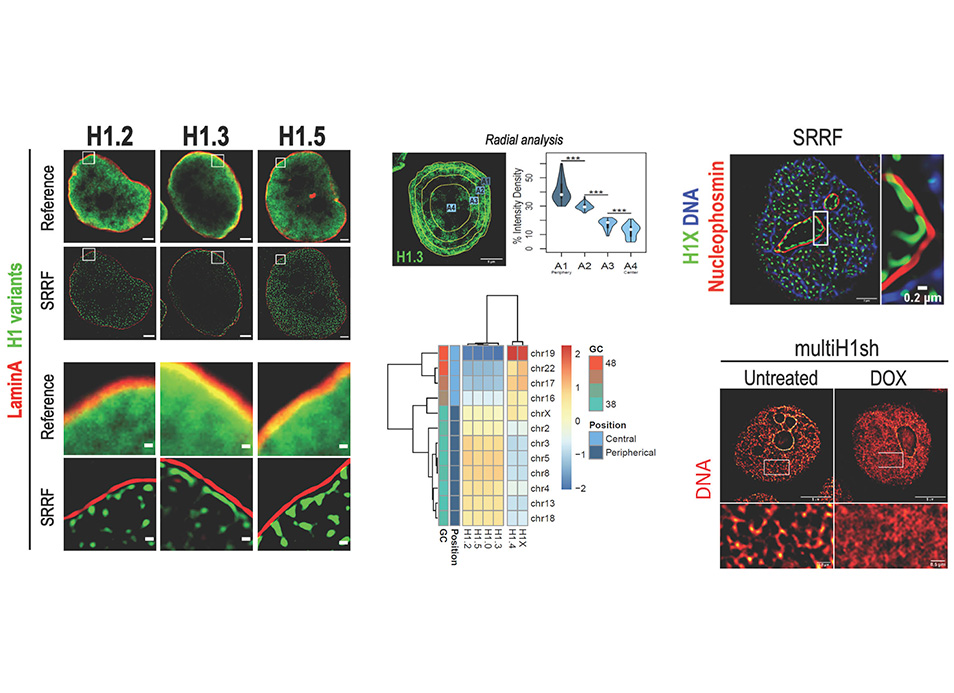
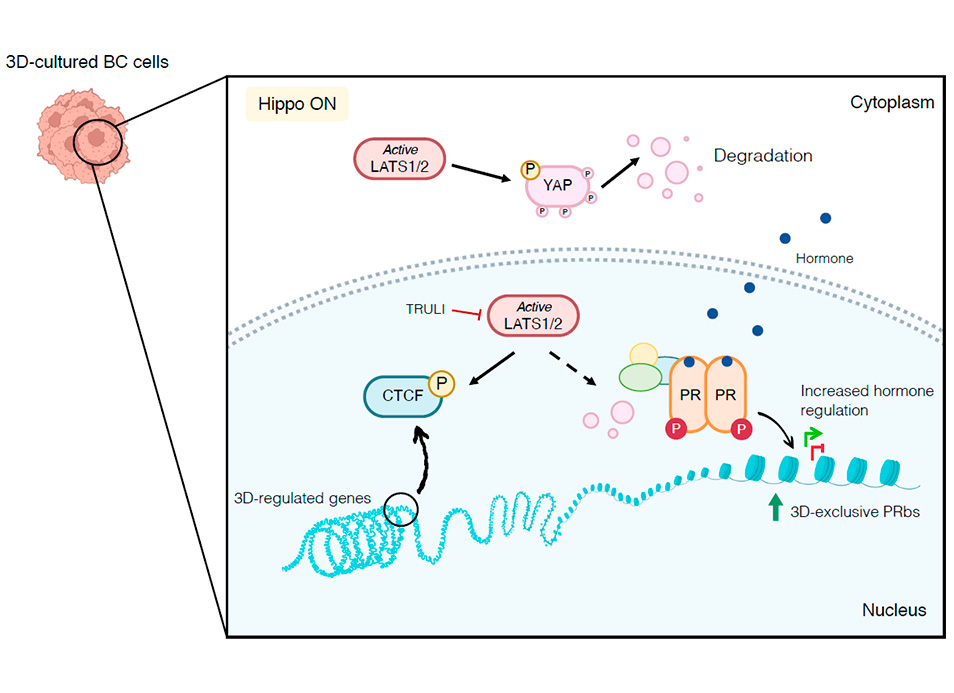
New publication in eLife of the Jordan Lab in collaboration with the IBMB Imaging Platform showing that…
An analysis of 60 metallopeptidase zymogens 3D structures revealed that their latency mechanism is less conserved than catalysis one.
The Proteolysis laboratory, directed by F. X. Gomis-Rüth, published a review about the latency of metallopeptidase zymogens at Chemical Reviews Journal, one of the most highly regarded and highest-ranked journals covering the general topic of chemistry.
This revision focuses on the structural determinants of inhibition in zymogens of metallopeptidases (MPs). These enzymes cleave polypeptides bound in the active-site cleft of catalytic domains through a general base/acid mechanism. This involves a solvent molecule bound to a catalytic zinc and general regulation of the mechanism through zymogen-based latency. Sixty reported structures from 11 metallopeptidase families reveal that prosegments, mostly N-terminal of the catalytic domain, block the cleft regardless of their size. Prosegments may be peptides (5−14 residues), which are only structured within the zymogens, or large moieties (<227 residues) of one or two folded domains. While some prosegments globally shield the catalytic domain through a few contacts, others specifically run across the cleft in the same or opposite direction as a substrate, making numerous interactions. Some prosegments block the zinc by replacing
the solvent with particular side chains, while others use terminal α-amino or carboxylate groups. Overall, metallopeptidase zymogens employ disparate mechanisms that diverge even within families, which supports that latency is less conserved than catalysis.