New publication in eLife of the Jordan Lab in collaboration with the IBMB Imaging Platform showing that…
De novo design of immunoglobulin-like domains
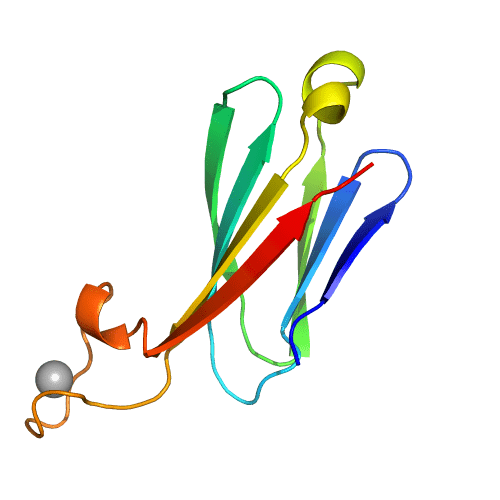
The design of immunoglobulin-like domains from scratch has been a long-standing challenge. We have identified principles and set up a computational approach for the de novo design of immunoglobulin-like domains with new structures, confirmed by X-ray crystallography and with the ability to scaffold ligand-binding loops.
Abstract
Antibodies and antibody derivatives such as nanobodies contain immunoglobulin-like (Ig) β-sandwich scaffolds which anchor the hypervariable antigen-binding loops and constitute the largest growing class of drugs. Current engineering strategies for this class of compounds rely on naturally existing Ig frameworks, which can be hard to modify and have limitations in manufacturability, designability and range of action. Here we develop design rules for the central feature of the Ig fold architecture – the non-local cross-β structure connecting the two β-sheets – and use these to de novo design highly stable Ig domains, confirm their structures through X-ray crystallography, and show they can correctly scaffold functional loops. Our approach opens the door to the design of antibody-like scaffolds with tailored structures and superior biophysical properties. This study has been carried out in collaboration between the Molecular Biology Institute of Barcelona (IBMB-CSIC) and the Institute for Protein Design at the University of Washington.
Reference:
Chidyausiku, T.M., Mendes, S.R., Klima, J.C., Gomis-Rüth, F. Xavier, Marcos, Enrique et al. De novo design of immunoglobulin-like domains. Nat Commun 13, 5661 (2022). https://doi.org/10.1038/s41467-022-33004-6